A filtration of the study region fibers according to minimal and maximal allowed length, and a separation into two groups depending on whether one or two of their ends are connected to the mesh.'
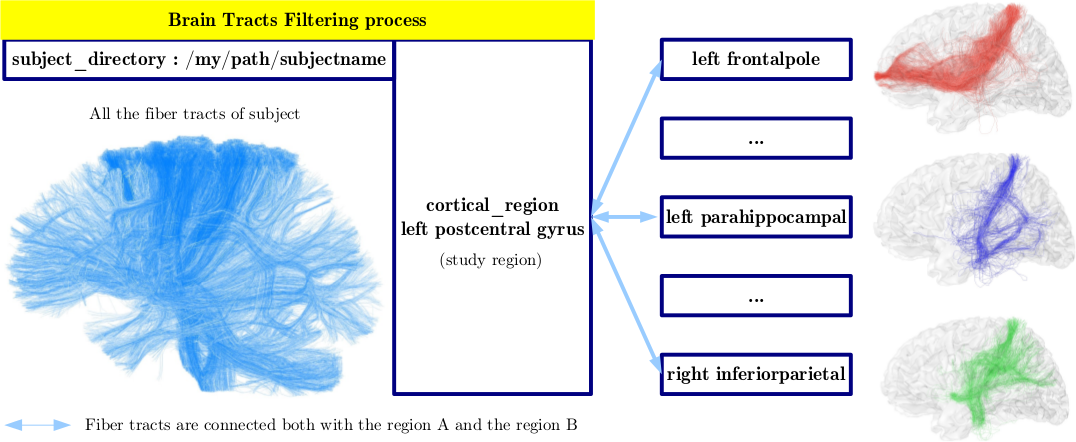
' Depending on the starting region, the fiber tracts are filtered according to three criteria:'
(1) only fibers connected to the starting region are retained (mandatory criteria)
(2) the fibers less than a threshold in length are removed (mandatory criteria)
(3) the fibers of longer than a threshold are removed (mandatory criteria)
This filtering is organized as follow:
- reads fibers: Only fibers generated by the Connectomist-2.0 and TrackVis softwares are accepted. It is possible to put a list of files.
- set gyrus pair name on each fiber: Each end of the fibers is associated with a vertex of the mesh, itself labeled by the starting region segmentation. This label is awarded to the fiber. If a fiber end is not identified on the mesh, it is labelled "notInMesh".
- select bundles from names: the fibers from at least one end to the starting region are kept.
- filter by length: smaller fibers than the threshold "minlength?" or larger than the threshold "maxlength_?" are deleted.
- merge the files: creating a single file output by fiber type (see below)
Two types of fiber tracts have to be taken into consideration in this process:
- labeled fibers are defined as having both ends attached to the mesh (and are consequently labelled)
- semilabeled fibers are defined as having only one end attached to the mesh (the other being not identified)
Usage example
regions_nomenclature: /casa/build/share/brainvisa-share-4.6/nomenclature/translation/nomenclature_desikan_freesurfer.txtoutputs_database: /my/path/brainvisa_dbstudy_name: studyAmethod: averaged approachregion: lh.inferiorparietalsubject_indir: /my/path/connectomist_db/B1500/StreamlineProbabilistic/aQBI/27seeds/001individual_white_mesh: /my/path/freesurfer_db/001/surf/bh.r.aims.white.giidw_to_t1: /my/path/connectomist_db/B1500/StreamlineProbabilistic/aQBI/27seeds/001/dw_to_t1.trmregions_parcellation: /my/path/freesurfer_db/group_analysis/average_group/average_brain/bh.annot.averagebrain.giifiber_tracts_format: bundlesmin_fibers_length: 20.0max_fibers_length: 500.0labeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/ default_tracking_session/connectivity_parcellation/avg/studyA/lh.inferiorparietal/filteredTracts/ 001_studyA_lh.inferiorparietal_labelled_fibers_20to500mm.bundlessemilabeled_fibers: /my/path/database_brainvisa/subjects/001/diffusion/default_acquisition/default_analysis/ default_tracking_session/connectivity_parcellation/avg/studyA/lh.inferiorparietal/filteredTracts/ 001_studyA_lh.inferiorparietal_semilabelled_fibers_20to500mm.bundles'
regions_nomenclature: Nomenclature ROIs File ( input )Nomenclature of the cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas
outputs_database: Choice ( input )This parameter retrieves all databases with brainvisa ontology present in your configuration, the generated files will be written on the chosen one (see the documentation to add a database in the BrainVisa configuration).
study_name: OpenChoice ( input )General name of the study.
method: Choice ( input )Two methods are proposed:
(1) averaged approach to obtain an average result on the group.
(2) concatenated approach to obtain individual results across the group.
region: OpenChoice ( input )The study region based on regions_nomenclature file.
subject_indir: Subject ( input )This is the subjects directory in a Connectomist database, where the fiber tracts files can be found.
individual_white_mesh: White Mesh ( input )Freesurfer white-grey interface of the cortex.
Should not be inflated.
dw_to_t1: Transform T2 Diffusion MR to Raw T1 MRI ( input )Affine spatial transformation to get the T1 MRI space from the dMRI diffusion (and tracts) space.
regions_parcellation: ROI Texture ( input )Cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas (?h.aparc.annot).
fiber_tracts_format: Choice ( input )Different fiber tracts formats are supported.
Selectable formats are:
(1) TrackVis file format with .trk as filename extension.
(2) Connectomist file format with .bundles as filename extension.
min_fibers_length: Float ( input )A filtering parameter to exclude low length fibers. Default to 20mm.
max_fibers_length: Float ( input )A filtering parameter to exclude long length fibers. Default to 500mm.
labeled_fibers: Filtered Fascicles Bundles ( output )All fibers having their ends identified on the mesh and corresponding to the various criteria in this process are merged into one output file.
semilabeled_fibers: Filtered Fascicles Bundles ( output )All fibers having only one end identified on the mesh and corresponding to the various criteria of this process are merged into one output file.
Toolbox : Constellation
User level : 2
Identifier :
constel_brain_tracts_filteringSupported file formats :
regions_nomenclature :Text file, Text filesubject_indir :Directory, Directoryindividual_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshdw_to_t1 :Transformation matrix, Transformation matrixregions_parcellation :GIFTI file, GIFTI file, Texturelabeled_fibers :Aims bundles, Aims bundlessemilabeled_fibers :Aims bundles, Aims bundles