Display a summary of the differents ways to compare reconstructions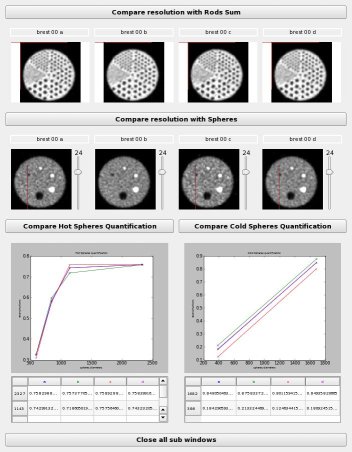
CT: Phantom CT ( input )
Pets_mean: ListOf( Phantom PET Mean ) ( input )
CT2PetMeans: ListOf( CT to PET transformation file ) ( input )
allRods: ListOf( Phantom Rods ) ( input )
allRods_sum: ListOf( Phantom Rods Sum ) ( input )
graph: Phantom Graph HotSph ( input )
graph_coldSph: Phantom Graph ColdSph ( input )
Toolbox : Nuclear Imaging
User level : 2
Identifier :
jzkDisplayQCFile name :
brainvisa/toolboxes/nuclearimaging/processes/jaszczak/jzkDisplayQC.pySupported file formats :
CT :NIFTI-1 image, NIFTI-1 imagePets_mean :NIFTI-1 image, NIFTI-1 imageCT2PetMeans :Transformation matrix, Transformation matrixallRods :NIFTI-1 image, NIFTI-1 imageallRods_sum :NIFTI-1 image, NIFTI-1 imagegraph :CSV file, CSV filegraph_coldSph :CSV file, CSV file