Segment spheres of the CT image allows you to find each center of spheres'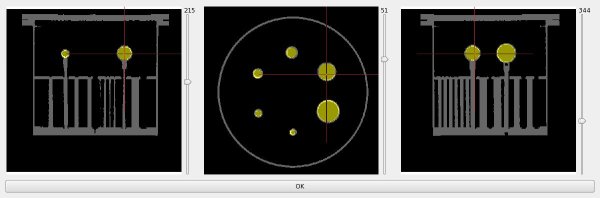
''First find Palette bounds using :
an approximative center of the biggest sphere (given by the user, or deduced thanks to the corresponding PET image) then launch ray from this approximative center to find a more precise center keep a sub volume of this biggest sphere, to have only the sphere (no sphere support) and finally thanks to the known volume of the biggest sphere, find the correct threshold Secondly find each sphere using :
palette bounds for threshold to apply to the Hough Algorithm. also, approximative sphere centers are used, but just like an optimisation for Hough Algorithm, selecting a subVolume bounding each sphere. Finally build an ROI for each sphere, thanks to the found centers and Jaszczak specifications.
'
run_parallele: Boolean ( input )
CT: Phantom CT ( input )
jzkSpec: Phantom Spec ( input )
sphdRaycast_max: ListOf( Integer ) ( input )
verbose: Boolean ( input )
Toolbox : Nuclear Imaging
User level : 2
Identifier :
jzkFindSphCenter_inCtFile name :
brainvisa/toolboxes/nuclearimaging/processes/jaszczak/jzkFindSphCenter_inCt.pySupported file formats :
CT :NIFTI-1 image, NIFTI-1 imagejzkSpec :CSV file, CSV file