All slices with rods are summed and then displayed, to check visually the resolution :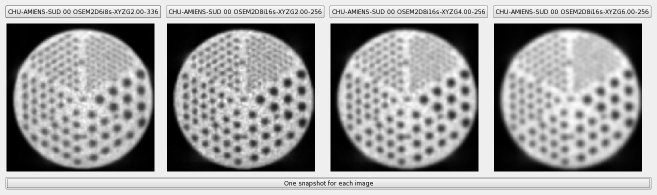
display: Choice ( input )
run_parallele: Boolean ( input )
CT: Phantom CT ( input )
Pets_mean: ListOf( Phantom PET Mean ) ( input )
CT2PetMeans: ListOf( CT to PET transformation file ) ( input )
jzkSpec: Phantom Spec ( input )
sumSize: Float ( optional, input )
percent: Float ( input )
allRods: ListOf( Phantom Rods ) ( output )
allRods_sum: ListOf( Phantom Rods Sum ) ( output )
verbose: Boolean ( input )
Toolbox : Nuclear Imaging
User level : 2
Identifier :
jzkSumRodsFile name :
brainvisa/toolboxes/nuclearimaging/processes/jaszczak/jzkSumRods.pySupported file formats :
CT :NIFTI-1 image, NIFTI-1 imagePets_mean :NIFTI-1 image, NIFTI-1 imageCT2PetMeans :Transformation matrix, Transformation matrixjzkSpec :CSV file, CSV fileallRods :NIFTI-1 image, NIFTI-1 imageallRods_sum :NIFTI-1 image, NIFTI-1 image