This process enables to calculate the bounding box parameters for each connected component using Voronoi diagram or directly from the mask.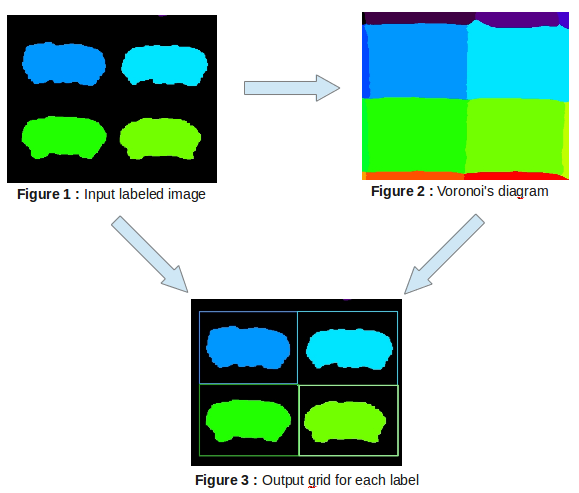
The bouding box calculation using Voronoi mode
Input_Volume_Label: 3D Volume ( input )Select the input labeled volume.
Voronoi_Mode: Boolean ( input )Enabling or disabling Vornoi mode.
Output_BoundingBox: Text file ( output )Enter the name of the output bounding box file.
Toolbox : Bioprocessing
User level : 3
Identifier :
ComputeBoundingBoxFromLabelFile name :
brainvisa/toolboxes/bioprocessing/processes/research/toolbox/ComputeBoundingBoxFromLabel.pySupported file formats :
Input_Volume_Label :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageOutput_BoundingBox :Minf, Minf