Parameterization of a sulcus, i.e. definition of a coordinate system on the sulcus. This is a way, amongst other things, to normalize sulci and compare them through individuals. As well as a parametrization, the process generates a text file that contains a depth curve and a sulcal profile that can be used for morphometrics purposes.
If one wants to reference such process, or look for detailed explanations about the methods, a few publications are mentioned below.
The parametrization process is described in:
Coulon O, Clouchoux C, Operto G, Dauchot K, Sirigu A, Anton J-L, "Cortical localization via surface parametrization : a sulcus-based approach", in Int. Conf. on Human Brain Mapping, Florence, Italy, 2006.
The use of the process for inter-subject comparison and depth curve statistics has been first presented in:
Cykowski MD, Coulon O, Kochunov PV, Amunts K, Lancaster JL, Laird AR, Glahn DC, Fox PT, "The central sulcus: an observer-independent characterization of sulcal landmarks and depth asymmetry", Cerebral Cortex, 18(9):1999-2009, 2008.
The sulcal profile and its potential use has been presented in:
Coulon O, Pizzagalli F, Operto G, Auzias G, Delon-Martin C, Dojat M, "Two new stable anatomical landmarks on the central sulcus: definition, automatic detection, and their relationship with primary motor functions of the hand", in IEEE EMBC 2011, 33rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp.7795-7798, Boston 2011.
The point is to define a coordinate system on a sulcus. This sulcus is identified by name or label in a graph. As an output, one gets a mesh of the sulcus, two coordinate textures, and one coordinate "grid" .
This process performs the parameterization of a sulcus. The sulcus needs to have been identified and labelled automatically or by hand before. The result is a x-y coordinate system on the mesh of the sulcus. Some geometrical features of the sulcus (such as the extremities or the top and bottome ridges) have constant cordinates across subjects and the parameterization therefore implicitely provides an inter-subject matching, which means that a particular coordinate pair (longitude, latitude) should localize the same anatomical point on the sulcus across subjects.
It is essential that the input graph contains proper information about the sulcus you want to parameterize. A graph that has been automatically labelled is likely NOT to contain a proper labelling. For instance the central sulcus will be labelled S.C.left. Sulcus being over-segmented, this label will point to every pieces of the central sulcus, including small branches you do not want, as shown by the figure below.
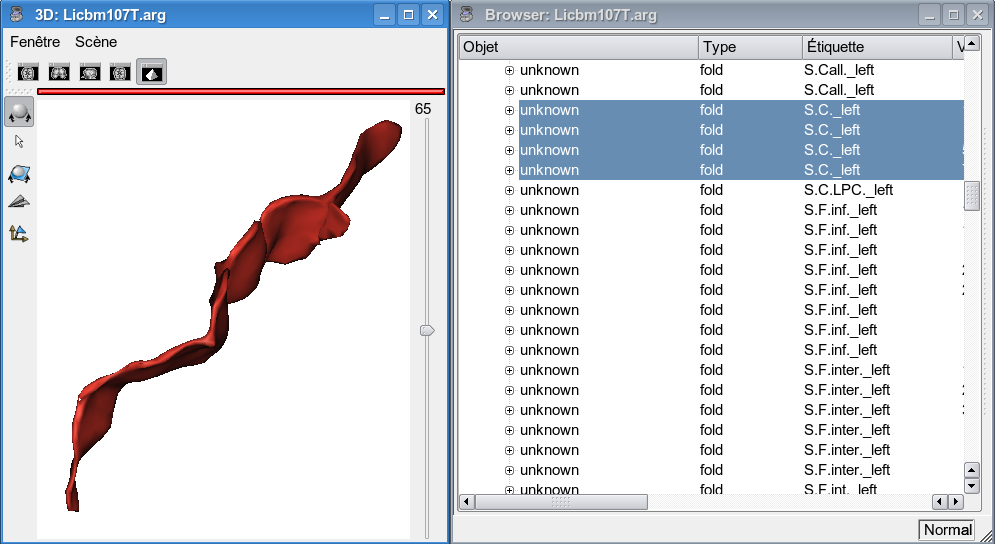
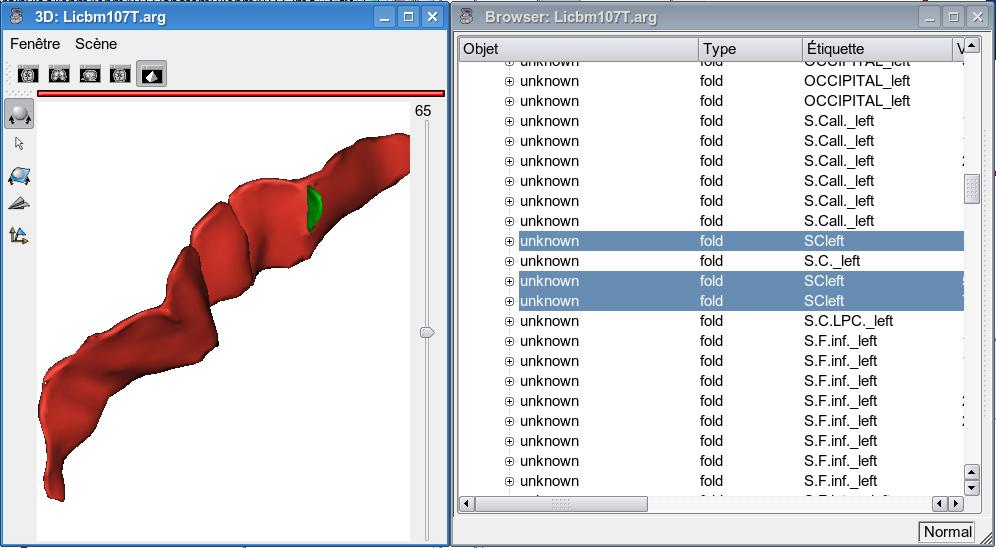
The solution is to display the graph (and a mesh) in an Anatomist 3D window, and the graph in an Anatomist browser. In the 3D window, select each pieces of the sulcus you want. They are highlighted in the browser, edit their label or name field with the identifier you want, e.g. scleft (see figure below), then save the graph. The best thing is to save the graph under a new name to avoid any corruption of a previous labelling. This new graph is the one to use for the sulcal parameterization.
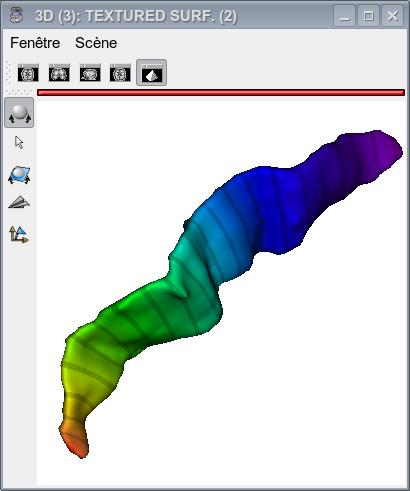
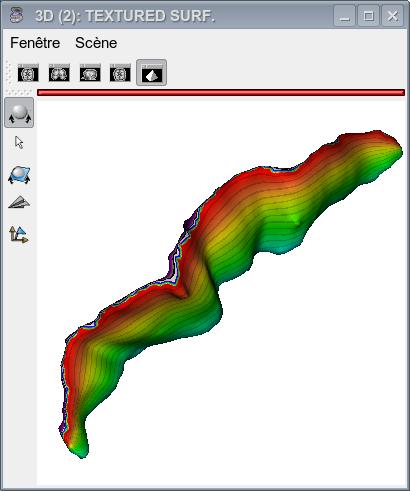
The figures below show the sulcus final mesh with the x-coordinate and y-coordinate textures as well as with the resulting coordinate grid.
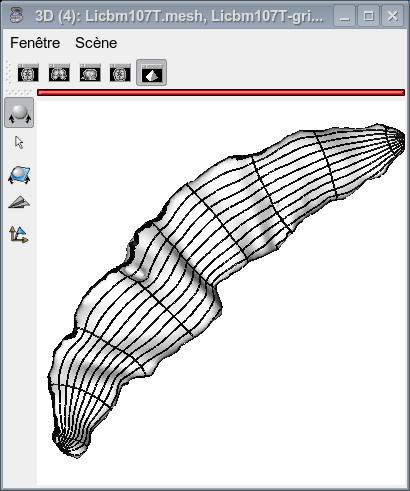
The process also returns a text file (depth_profile). This file contains 3 colums. The first one is a list of 101 positions (from 0 to 100), the second one is the depth curve value for each of these positions, and the third one is the sulcal profile value for eachof these positions.
A detailed documentation is available on Olivier's page (if the link does not work, specify your browser path in the preferences and restart Brainvisa).
graph: Cortical folds graph ( input )Input sulci graph
mri: T1 MRI Bias Corrected ( input )subject's T1 MRI
label_attributes: Choice ( input )The sulcus can be identified by name or label
label_values: String ( input )value of the name/label to extract the sulcus
orientation: Choice ( input )Main orientatiuon of the sulcus
sulcus_mesh: Sulcus mesh ( output )output mesh: the sulcus is remeshed as a single object
texture_param1: Sulcus x coordinate texture ( output )1rst parameter texture (x)
texture_param2: Sulcus y coordinate texture ( output )2nd parameter texture (y)
coordinates_grid: Sulcus coordinate grid mesh ( output )Mesh of the coordinate grid (to be uses for visualization only)
depth_profile: Sulcus depth profile ( output )The file that contains the depth curve and sulcal profile
dilation: Float ( input )Dilation parameter. Keep the default value.
morpho_offset: Choice ( input )
meshDecimation: Boolean ( input )
deltaT: Float ( input )
stop: Float ( input )
Toolbox : Cortical Surface
User level : 0
Identifier :
ParameterizeSulcusFile name :
brainvisa/toolboxes/cortical_surface/processes/anatomy/obsolete/ParameterizeSulcus.pySupported file formats :
graph :Graph and data, Graph and datamri :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesulcus_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshtexture_param1 :Texture, Texturetexture_param2 :Texture, Texturecoordinates_grid :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshdepth_profile :Text file, Text file