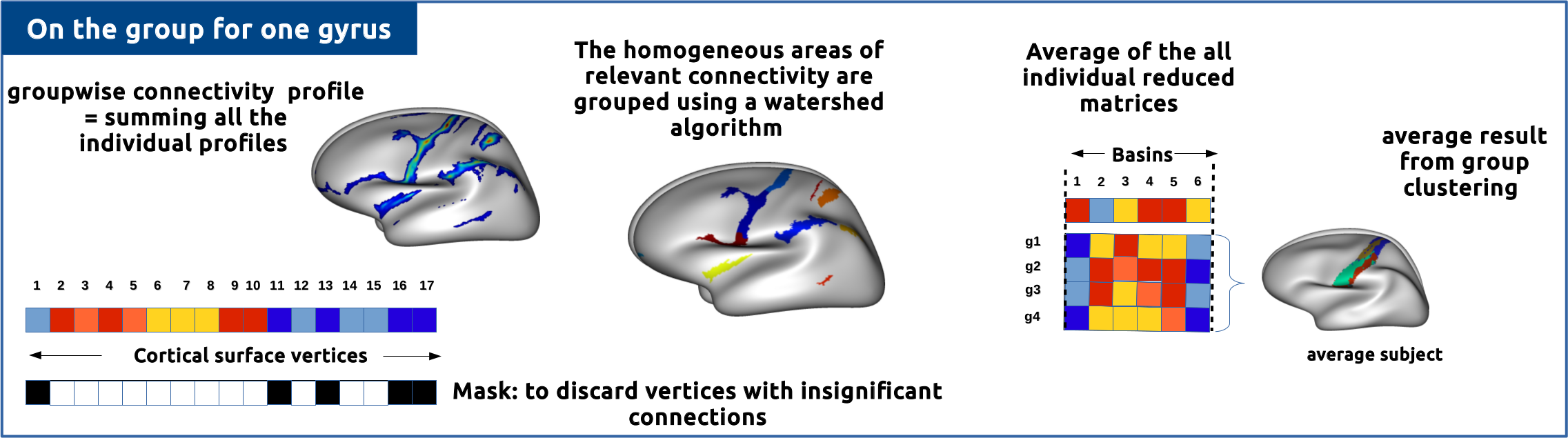
Provides a clustering of a group of subjects.
Generalities
Method
Usage example
regions_nomenclature: /my/path/share/brainvisa-share-4.6/nomenclature/translation/nomenclature_desikan_freesurfer.txtstudy_name: studyAnew_study_name: group_studyAmethod: averaged approachregion: lh.inferiorparietalsubjects_group: /my/path/brainvisa_db/subjects/group_analysis/g1/g1_group.xmlmean_individual_profiles: '/my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/ initiation/lh.inferiorparietal/clustering/smooth3.0/001_studyA_lh.inferiorparietal_mean_profile_20to500mm.gii'normed_individual_profiles: '/my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/ initiation/lh.inferiorparietal/clustering/smooth3.0/001_studyA_lh.inferiorparietal_normed_mean_profile_20to500mm.gii'average_mesh: /my/path/freesurfer_db/group_analysis/average_mesh_group/average_brain/averagebrain.white.giiregions_parcellation: '/my/path/freesurfer_db/group_analysis/average_mesh_group/average_brain/bh.annot.averagebrain.gii'smoothing: 3.0nb_clusters: 12
regions_nomenclature: Nomenclature ROIs File ( input )Nomenclature of the cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas
study_name: OpenChoice ( input )General name of the study, links to corresponding study data in output_database to retrieve profiles computed during the individual processing.
new_study_name: String ( optional, input )Optional name given to the group study.
Default to study_name.
method: Choice ( input )Two methods are proposed:
(1) averaged approach to obtain an average result on the group.
(2) concatenated approach to obtain individual results across the group.
region: OpenChoice ( input )The study region based on regions_nomenclature file.
subjects_group: Group definition ( input )
mean_individual_profiles: ListOf( Connectivity Profile Texture ) ( input )List of the mean individual profiles of the given group of subjects.
normed_individual_profiles: ListOf( Connectivity Profile Texture ) ( input )List of the normed individual profiles of the given group of subjects.
average_mesh: White Mesh ( input )Freesurfer average white mesh of a group of subjects.
regions_parcellation: ListOf( ROI Texture ) ( input )Cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas (?h.aparc.annot).
Implications of the choice of the method on the regions_parcellation parameter:
(1) averaged approach: the regions_parcellation parameter must be a file representing the average parcellation of the group of subjects.
(2) concatenated approach: the regions_parcellation parameter must be a list of files, each representing the individual parcellation of a subject of the group.
smoothing: Float ( input )Degree of smoothing (in millimetres).
Default to 3.0 mm.
nb_clusters: Integer ( input )Maximal number of clusters used to parcellate the study region.
Default to 12.
Toolbox : Constellation
User level : 1
Identifier :
constel_group_pipelineFile name :
brainvisa/toolboxes/constellation/processes/group_pipeline/constel_group_pipeline.pySupported file formats :
regions_nomenclature :Text file, Text filesubjects_group :XML, XMLmean_individual_profiles :GIFTI file, GIFTI file, Texturenormed_individual_profiles :GIFTI file, GIFTI file, Textureaverage_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshregions_parcellation :GIFTI file, GIFTI file, Texture