Calculation of the mean individual profile by summing all vertices of the cortical region.
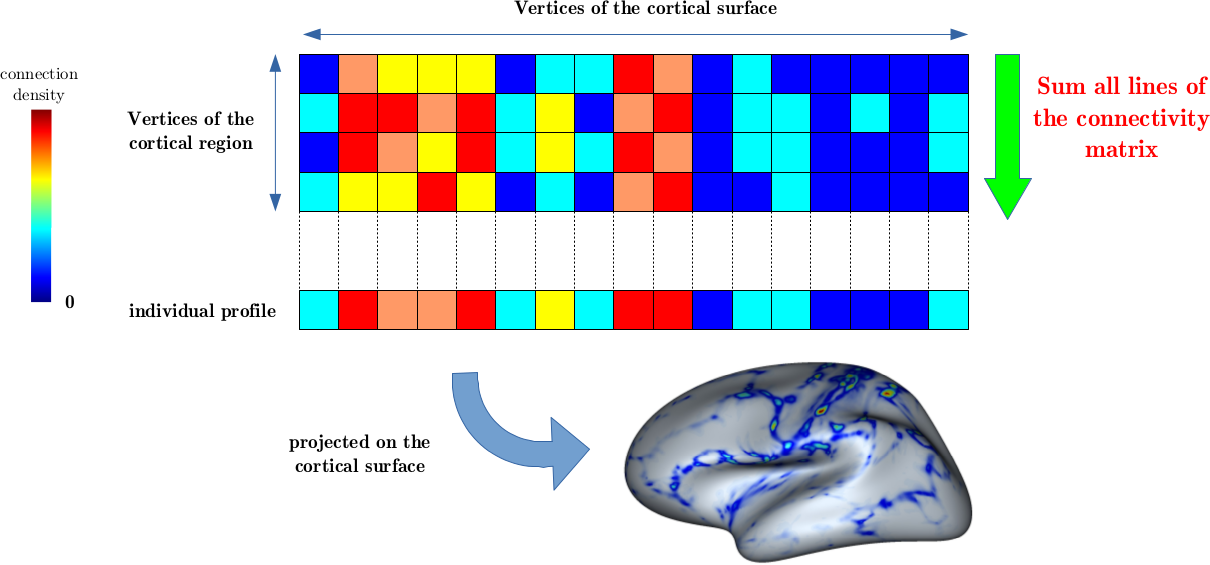
In each matrix, the ith line corresponds to the cortical connectivity profile of the ith point of the patch, representing its connexions towards each point of the cortical surface. Then, all profiles are averaged across all patch points. So, the connectivity profile of the patch is computed.
Usage example
regions_nomenclature: /casa/build/share/brainvisa-share-4.6/nomenclature/translation/nomenclature_desikan_freesurfer.txtregion: lh.inferiorparietalcomplete_matrix_smoothed: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation /avg/studyA/lh.inferiorparietal/matrix/001_studyA_lh.inferiorparietal_complete_matrix_smooth3.0_20.0to500.0mm.imasregions_parcellation: /my/path/freesurfer_db/group_analysis/average_group/average_brain/bh.annot.averagebrain.giimean_individual_profile: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation /avg/studyA/lh.inferiorparietal/clustering/smooth3.0/001_studyA_lh.inferiorparietal_mean_profile_20.0to500.0mm.giierase_matrix: True
regions_nomenclature: Nomenclature ROIs File ( input )Nomenclature of the cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas
region: String ( input )The study region based on regions_nomenclature file.
complete_matrix_smoothed: Connectivity Matrix ( input )Smoothed connectivity matrix.
regions_parcellation: ROI Texture ( input )Cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas (?h.aparc.annot).
erase_matrices: Boolean ( input )
mean_individual_profile: Connectivity Profile Texture ( output )Mean individual profile calculated from smoothed matrix.
Toolbox : Constellation
User level : 2
Identifier :
constel_mean_individual_profileSupported file formats :
regions_nomenclature :Text file, Text filecomplete_matrix_smoothed :Sparse Matrix, Sparse Matrixregions_parcellation :GIFTI file, GIFTI file, Texturemean_individual_profile :GIFTI file, GIFTI file, Texture