Construction of the cortical region connectivity matrix.
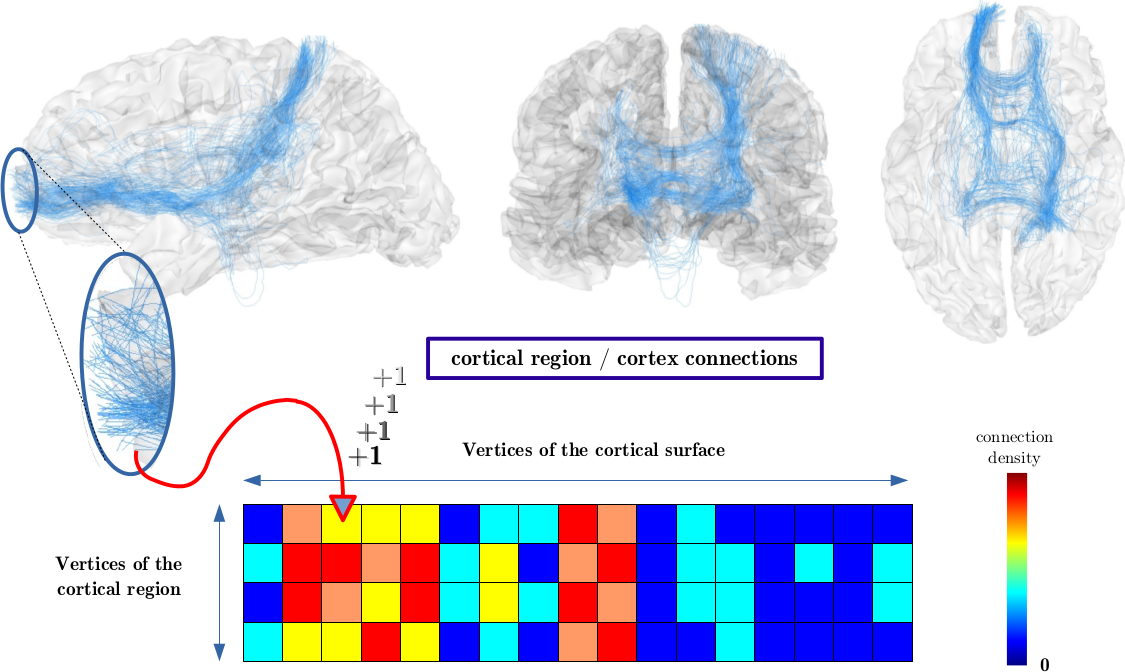
(1) Each fibertract connects two vertices of the cortical surface and contributes to the cortical connectivity matrix.
(2) The matrix where each line corresponds to the connectivity profile of one vertex of the input cortical region with the whole cortical surface.
Usage example
regions_nomenclature: /casa/build/share/brainvisa-share-4.6/nomenclature/translation/nomenclature_desikan_freesurfer.txtregion: lh.inferiorparietaloversampled_semilabeled_fibers: /my/path/database_brainvisa/subjects/001/diffusion/default_acquisition/default_analysis/ default_tracking_session/ connectivity_parcellation/avg/studyA/lh.inferiorparietal/filteredTracts/ 001_studyA_lh.inferiorparietal_oversampled_semilabelled_fibers_20to500mm.bundleslabeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/ default_tracking_session/connectivity_parcellation/avg/studyA/lh.inferiorparietal/filteredTracts/ 001_studyA_lh.inferiorparietal_labelled_fibers_20to500mm.bundlesindividual_white_mesh: /my/path/freesurfer_db/001/surf/bh.r.aims.white.giidw_to_t1: /my/path/connectomist_db/B1500/StreamlineProbabilistic/aQBI/27seeds/001/dw_to_t1.trmregions_parcellation: /my/path/freesurfer_db/group_analysis/average_group/average_brain/bh.annot.averagebrain.giimatrix_semilabeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/studyA/ lh.inferiorparietal/matrix/001_studyA_lh.inferiorparietal_semilabelled_fibers_matrix_20to500mm.imasmatrix_labeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/studyA/ lh.inferiorparietal/matrix/001_studyA_lh.inferiorparietal_labelled_fibers_matrix_20to500mm.imasprofile_semilabeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/studyA/ lh.inferiorparietal/matrix/001_studyA_lh.inferiorparietal_semilabelled_fibers_mean_profile_20to500mm.imasprofile_labeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/studyA/ lh.inferiorparietal/matrix/001_studyA_lh.inferiorparietal_labelled_fibers_meanprofile_20to500mm.imascomplete_individual_matrix: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation/avg/studyA/ lh.inferiorparietal/matrix/001_studyA_lh.inferiorparietal_complete_matrix_smooth0.0_20to500mm.imas
regions_nomenclature: Nomenclature ROIs File ( input )Nomenclature of the cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas
region: String ( input )The study region based on regions_nomenclature file.
oversampled_semilabeled_fibers: Filtered Fascicles Bundles ( input )All the fibers having one end identified on the mesh and oversampled.
labeled_fibers: Filtered Fascicles Bundles ( input )All the fibers having their ends identified on the mesh.
individual_white_mesh: White Mesh ( input )Freesurfer white-grey interface of the cortex.
Should not be inflated.
dw_to_t1: Transform T2 Diffusion MR to Raw T1 MRI ( input )Affine spatial transformation to get the T1 MRI space from the dMRI diffusion (and tracts) space.
regions_parcellation: ROI Texture ( input )Cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas (?h.aparc.annot).
matrix_semilabeled_fibers: Connectivity Matrix ( output )Connectivity matrix that links labeled vertices to unlabeled vertices on the given cortical parcellation.
matrix_labeled_fibers: Connectivity Matrix ( output )Connectivity matrix that links labeled vertices of the study region to other labeled vertices of the cortex on the given cortical parcellation.
profile_semilabeled_fibers: Connectivity Profile Texture ( output )Projection of the semilabeled connectivity matrix on the cortical surface.
profile_labeled_fibers: Connectivity Profile Texture ( output )Projection of the labeled connectivity matrix on the cortical surface.
complete_individual_matrix: Connectivity Matrix ( output )Superposition of both semilabeled and labeled matrices.
Toolbox : Constellation
User level : 2
Identifier :
constel_sparse_individual_matricesSupported file formats :
regions_nomenclature :Text file, Text fileoversampled_semilabeled_fibers :Aims bundles, Aims bundles, Trackvis tractslabeled_fibers :Aims bundles, Aims bundles, Trackvis tractsindividual_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshdw_to_t1 :Transformation matrix, Transformation matrixregions_parcellation :GIFTI file, GIFTI file, Texturematrix_semilabeled_fibers :Sparse Matrix, Sparse Matrixmatrix_labeled_fibers :Sparse Matrix, Sparse Matrixprofile_semilabeled_fibers :GIFTI file, GIFTI file, Textureprofile_labeled_fibers :GIFTI file, GIFTI file, Texturecomplete_individual_matrix :Sparse Matrix, Sparse Matrix