Selection of commissures
To rely on the organisation provided by Brainvisa when using the segmentation softwares of SHFJ, you should trigger this preprocessing tool, which allows the selection of the intersection between the commissures and the interhemispheric plane, and one additional interhemispheric plane point, in order to compute the transformation towards Talairach's reference system. This referential allows various checking and initialization (but images are not resampled). If your image has been normalized using SPM or MRItotal (MNI tool), you can specify it to get rid of the manual selection (check with one example if this short cut works for you).
To use that tool for one given subject, you must have previously imported the T1 MRI of the subject into Brainvisa's "database": to do this, use the anatomical MRI importation process.
To check that Brainvisa has understood the existence of this new subject, open the treatment "Prepare Subject for Anatomical Pipeline".
Click on the magnifying glass to get access to the list of subjects registered in the Brainvisa database.
Your new subject should be there. If your database includes many subjects, you can trigger sorting operations with "protocole, voxel_size..." buttons.
You have found your subject name, select it:
The eye of Brainvisa is now active, which means the MR scan can be sent to Anatomist visualisation software.
Click on this eye...
You should see now the following windows:
You now have to specify the commissure localisations. If your image has been spatially normalised using one of the procedures registered in Brainvisa, you just have to make your choice in the "Normalised" menu. Then execute the treatment...
Registered procedures are:
Warning: you have to be aware that if your image has not been resampled to a high spatial resolution, the segmentation tools will give bad results, even if the commissures are manually selected... The spatial resolution of cortical fold, indeed, is often around 1mm. Furthermore, if your initial data had a large slice thickness, you will have a lot of problems...
- SHFJ from SPM: minimal field of view, resolution 1x1x1mm
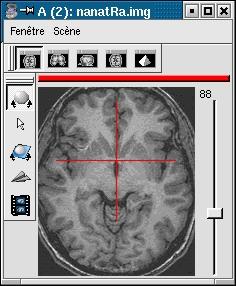
- MNI from Mritotal: resolution 1x1x1mm
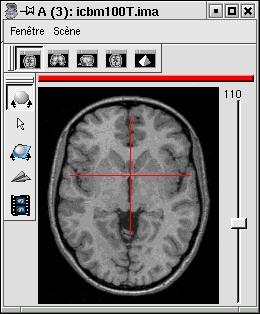
- Marseille from SPM: resolution 1x1x1mm but larger field of view
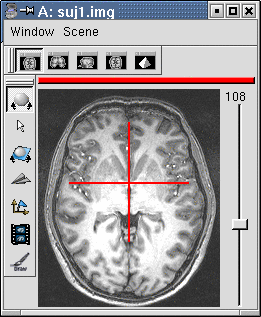
NB: It is not necessary to get a perfect selection. Automatic spatial normalisation procedures, indeed, do not take into account the exact localisation of the commissures... If you are not completely sure of you, check the ASCII file "mri/anubis.APC" created by Brainvisa (you can click on the commissure eye to put it in "nedit" editor). If all seems to work, you can iterate this treatment on your whole normalized subject set.
If your image is not spatially normalised, Anatomist will help you to click on the 3 or 4 points of interest.
- If the volume is not already visible, click on the edit button of one of the point parameters to have Anatomist display it
- Set the Anatomist view so as to see the point (use the slice slider) and click in it on the wished point.
- When the cursor position is good, click on the button of the 3D point in BrainVISA: the position is transfered from Anatomist to BrainVISA.
- If you are not happy with the position, you can restart the operation: click again on other points then on the parameter button to validate: a new position replaces the previous one in BrainVisa.
You have to select 3 points (you can use a sagittal slice if you prefer):
- Intersection of anterior commissure with interhemispheric plane;
- Intersection of posterior commissure with interhemispheric plane;
- Any point of the interhemispheric plane not aligned with the previous ones.
- Optionally, any point of the left hemisphere. Specifying this point is not mandatory, it is only useful to detect a possible orientation problem on the input image. Be careful, axial and coronal views in Anatomist are in radiological convention by default (left and right are flipped).
NB: if the colormap is not perfectly tuned, you can have access to it. Select your MR scan in Anatomist control window, and look for the colormap in the "object-specific" menu.
Anterior commissure:
Posterior commissure:
Inter-hemispheric plane:
Point of the left hemisphere (on the right on axial slices displayed in radiological convention):
You should now have filled the 12 coordinate fields:
A last parameter, allow_flip_initial_MRI, specifies if you allow or forbid BrainVisa to re-write the input image if it happens to need changing orientation before going through the segmentation pipeline. Indeed our tools assume that images are in axial orientation and are in a radiological and indirect referential. If they are not, hemispheres separation is likely to fail, sulci identification will not be able to produce sane results, and the link to a common referential (Talairach) will be wrong.
Execute the treatment... It will create the ASCII file anubis.APC, which you can visualise with a second Brainvisa eye.
NB: SHFJ segmentation tools can be used without that file, but with a lesser robustness. Anyway, for the sake of security, we do not allow you to trigger a pipeline as long as this file does not exist...
T1mri: Raw T1 MRI ( input )
commissure_coordinates: Commissure coordinates ( output )
Normalised: Choice ( input )Spatial normalisation procedure
Anterior_Commissure: Point3D ( optional, input )Position of the anterior commissure (mm)
Posterior_Commissure: Point3D ( optional, input )PC (mm)
Interhemispheric_Point: Point3D ( optional, input )A point of the inter-hemispheric plane (mm). Take it as far as possible from AC and PC (for instance in the upper part of the brain)
Left_Hemisphere_Point: Point3D ( optional, input )Any point of the left hemisphere (not too close to the inter-hemispheric plane)
allow_flip_initial_MRI: Boolean ( input )Allows or forbids re-writing the original image after flipping it if needed
reoriented_t1mri: Raw T1 MRI ( output )
remove_older_MNI_normalization: Boolean ( input )This parameters triggers the removal of older normalization information of the same data in the MNI space, to prevent ambiguities between contradictory transformations.
older_MNI_normalization: Transform Raw T1 MRI to Talairach-MNI template-SPM ( optional, input )Older normalization information going to the MNI space. It will just be removed, if present, and if remove_older_MNI_normalization is set to True.
Toolbox : Morphologist
User level : 0
Identifier :
preparesubjectFile name :
brainvisa/toolboxes/morphologist/processes/segmentationpipeline/components/preparesubject.pySupported file formats :
T1mri :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagecommissure_coordinates :Commissure coordinates, Commissure coordinatesreoriented_t1mri :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageolder_MNI_normalization :Transformation matrix, Transformation matrix