This pipeline performs a bayesian segmentation of MR images, with some parameters optimized by Expectation-Maximization (noise, bias, Gaussian mixture...).
The robustness of the process is increased by initializing mixture parameters from a naive 4-class mixture.
Several preprocessing steps for the non-stationary prior probabilities are possible. If no probabilisitic prior is available, it can be derived from "hard" labels.
Pipeline steps
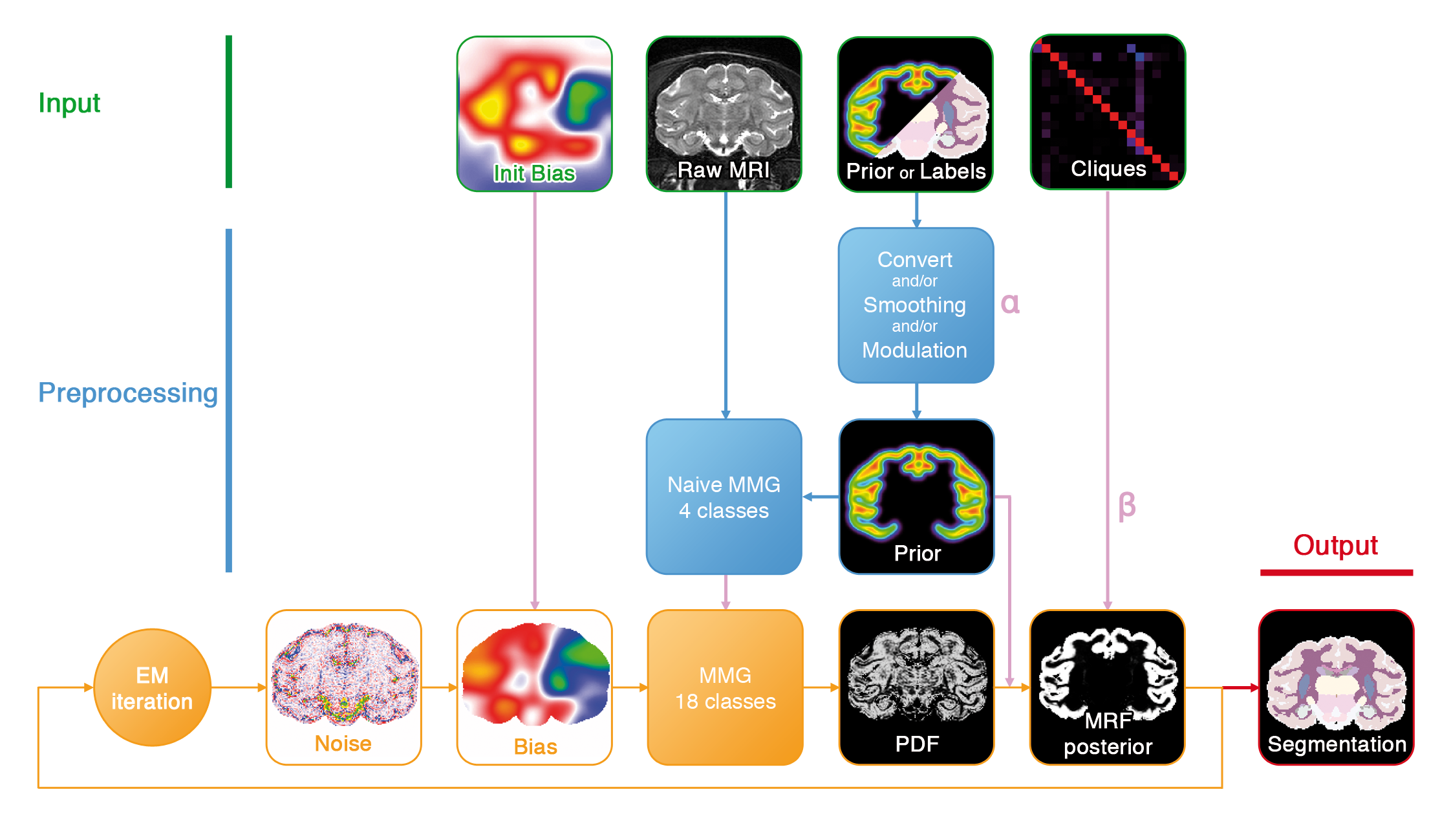
Figure 1. Segmentation pipeline.
- Mixture Model Initialization: probabilizes the atlas if needed and initializes the GMM parameters.
- EM Segmentation: fits a statistical model to the MRI by expectation-maximization. Intensities are assumed to originate from a gaussian mixture model on a MRF lattice with non-stationary priors. This step also includes densoising and bias field estimation .
analysis: String ( optional, input )Name of the analysis attribute that should be attached to this pipeline output files. If not provided, decision is let to the auto filling system.
input_mri: P:MRI Raw ( input )Raw MRI.
specie: OpenChoice ( optional, input )
contrast: OpenChoice ( optional, input )
is_mapping: Boolean ( optional, input )Set to true if the input image is a parametric map (T1 mapping, T2 mapping, etc) instead of a magnitude T1 or T2 weighted image.
mask: P:Atlas Registered Skull Stripping Mask ( optional, input )Skull-stripping mask. All subsequent analyses will be restricted to voxels inside this mask (parameter estimation, segmentation...). It will also be used to lower the memory load by only loading data inside the mask bounding box.
bias_field: P:MRI VIP Bias Field ( optional, input )Bias field initialization. If provided, the bias field will be initialized with this image before EM refinement.
labels: P:Atlas Registered Labels ( optional, input )Labels registered into the input image space. If no truely probabilitic prior is abailable, they can be used to derive prior probabilities by converting them to a 4D image and smoothing it with a Gaussian kernel.
prior: P:Atlas Registered Prior ( optional, input )Non-stationary prior probabilities. This 4D volume should contain a priori probabilities to observe a given class at a any location.
clique_matrix: P:Atlas Clique Matrix ( optional, input )Matrix containing a priori probabilities on cliques, i.e. possible neighbouring configurations.
hierarchy: P:Atlas Labels Hierarchy ( optional, input )Hierarchy associated with the atlas used (probabilistic priors or registered labels).
graywhite_hierarchy: P:Atlas Grey/White Hierarchy ( optional, input )A hierarchy file that classifies each label in one of 5 naive classes (background, CSF, white matter, gray matter, white-gray mixture). It is used to convert the naive 4-class mixture into a full anatomical mixture.
output_prior: P:EM Prior Proba ( optional, output )Non-stationary prior used as input for the bayesian segmentation. It can be the unchanged input prior, labels converted to probabilities, or a modulated version of these two possibilities.
output_posterior: P:EM Parcellation Proba ( optional, output )Output posterior probabilities, after EM optimization of the model parameters.
output_labels: P:EM Parcellation ( optional, output )Output segmentation.
Toolbox : Primatologist
User level : 0
Identifier :
primate_SegmentationPipelineFile name :
brainvisa/toolboxes/primatologist/processes/pipelines/primate_SegmentationPipeline.pySupported file formats :
input_mri :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagemask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagebias_field :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagelabels :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageprior :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageclique_matrix :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehierarchy :Hierarchy, Hierarchygraywhite_hierarchy :Hierarchy, Hierarchyoutput_prior :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageoutput_posterior :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageoutput_labels :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 image