Display connectivity from the connectivity matrix
Display full (non-reduced) connectivity matrix information for a given patch (gyrus).
For a viewer using a reduced matrix, see Anatomist show connectivity profiles of a specific cortical element.Connectivity from a point of the patch
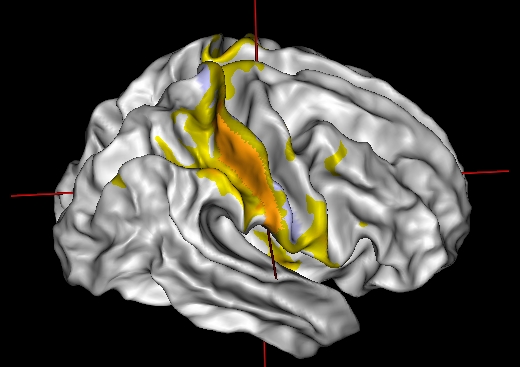
Simple left-click on the patch in Anatomist view
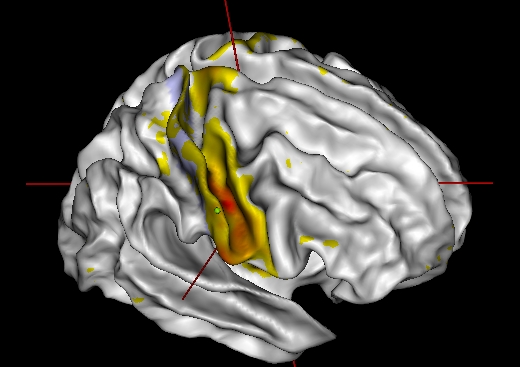
Connectivity to a point outside the patch
Left-click on a point of the cortex outside the patch
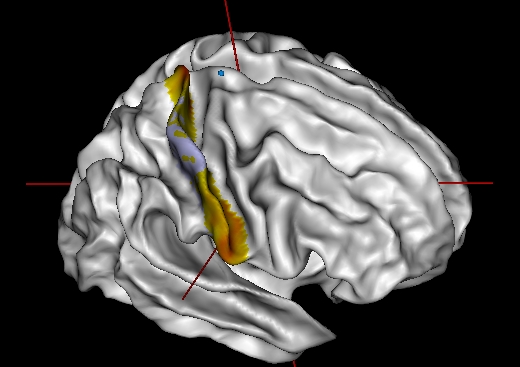
connectivity of a cluster within the patch
This mode only works if the gyrus_texture parameter is a clustering texture of the patch. It may have several timesteps (several numbers of clusters).
Ctrl + left-click on a point in the patch.
connectivity_matrix: Matrice de connectivité ( entrée )
white_mesh: White Mesh ( entrée )
gyrus_texture: ROI Texture ( entrée )
Toolbox : Constellation
Niveau d'utilisateur : 2
Identifiant :
anatomistViewConnmatrixNom de fichier :
brainvisa/toolboxes/constellation/processes/viewers/anatomistViewConnmatrix.pySupported file formats :
connectivity_matrix :gz compressed NIFTI-1 image, GIS image, NIFTI-1 image, Sparse Matrix, gz compressed NIFTI-1 imagewhite_mesh :GIFTI file, GIFTI file, Maillage MESH, MNI OBJ mesh, PLY mesh, Maillage TRI, Z compressed GIFTI file, Z compressed GIFTI file, Z compressed MESH mesh, Z compressed MNI OBJ mesh, Z compressed PLY mesh, Z compressed TRI mesh, gz compressed GIFTI file, gz compressed GIFTI file, gz compressed MESH mesh, gz compressed MNI OBJ mesh, gz compressed PLY mesh, gz compressed TRI meshgyrus_texture :GIFTI file, GIFTI file, Texture, Z compressed GIFTI file, Z compressed GIFTI file, Z compressed Texture, gz compressed GIFTI file, gz compressed GIFTI file, gz compressed Texture