Guide to overcome Ana Brain Mask From T1 MRI problems
For some reason, the procedure Brain Mask From T1 MRI failed for one of your images. A solution may be provided by this procedure, which proposes variants of the standard one. You can choose one of these variants in the menu. After applying this treatment, the result can be visualized in Anatomist if you select Mask visualization.
If nothing solve your problems, you will have to perform some manual corrections with Anatomist drawing's capacities: Label volume editor (you can use a shortcut to this treatment through a dedicated icon in brainVISA interface).
If you have no real understanding of the process leading to extract the brain mask, just try the variants one after another until you reach a good result. If none of the variants is successful, you can get access to more parameters either in brainVISA using Vip Get Brain, or from the command line (VipGetBrain -help). Finally, it should be possible to perform a manual correction with Anatomist's drawing module, but no documentation is provided yet.
extra-cerebral structures
The mask includes anatomical structures which do not belong to the brain:
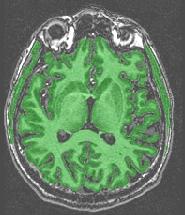
This is a failure of the erosion process supposed to break down brain connexions with the outside world (optical nerves, etc.). This failure results from a bad tuning of the erosion size during the automatic adjustment loop. Usually, the extra-cerebral structures are connected to cerebellum and/or temporal lobes. If only the cerebellum has this problem, you can proceed further because the cerebellum will be removed during further processing. Otherwise, try one of the variants with fixed erosion size, trying to keep this size as small as possible. In case of success, check that the erosion did not become too large. In such a case, some gyri may have been deleted. Then, you will have to choose the best trade-off.
If the previous choices did not work, the problem may stem from some artifact hiding in some places the black areas corresponding to the skull:
In such cases, the initial binarization which is trying to fill small cavities resulting from noise has to be discarded, otherwise the skull is partly cancelled and the erosion has not a chance to do its job. Try the following variants:
"Standard/Robust... without regularisation"
Another source of trouble is MR reconstruction artifacts in the first/last slices. You can blank some of these slices using two parameters: first_slice and last_slice. Then try the standard procedure or one of its variants:
"Standard (iterative erosion from 2mm)"
Eroded cerebral structures
Some of the gyri have been lost somewhere:
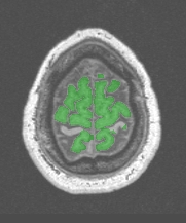
This problem occurs with very thin gyri, which width is less than two times the erosion size. Try the variants beginning with a 1.5mm erosion size:
"Standard/Robust (iterative or fixed erosion from 1.5mm)"
If it is not sufficient, try the procedure "Vip Get Brain" or the command line VipGetBrain, following these lines:Do not be too maniac, these eroded gyri may be forget for most applications.
- Try to reduce erosion size;
- Check histogram analysis, manually lower grey matter mean or increase grey matter standard deviation;
- Increase the number of iterations in the classification regularization.
Strange results
the numerous refinements of the simple erosion/dilation procedure allowing robustness to a wide range of images may sometime create strange problems. For instance, with "Robust..." procedures, a piece of white matter may be deleted in corpus callosum and brain stem:
The best solution is the use of the simplest variants:
"Fast (2mm erosion)"
"Fast (2.5mm erosion)"
"Fast (3mm erosion)"
Failure of histogram analysis
To perform the initial binarization, namely to create the black and white image describing the range of tissues which may belong to the brain, an automatic analysis of the 3D image's histogram is computed. This analysis is looking for two modes/peaks corresponding to grey and white matter (Vip Histogram Analysis). This analysis is successful if two cyan lines stem from these two peaks, like in the following example:
You may have access to this visualization (gnuplot required), if you choose"Histogram analysis visualization"
in the help menu. (choose Nothing in the variant menu). Unfortunatelly, this choise freezes the brainVISA procedure (stupid technical problem), hence you should not use it without good understanding of what you loose...
If your analysis is not similar to the previous example, the problem may stem from a failure of the bias correction procedure (Other examples there). In such a case try to modify the bias correction options.
If your image suffers from a lack of grey/white contrast (monkey images, bad coil, etc.), you can provide manually the grey/white statistics, using any text editor (textedit,nedit,xemacs,vi,etc...). If your computer provides nedit, you have access to the file *.han (located in anatomy directory), with brainVISA's eye. The file format is the following:
sequence: inversion recovery
background: mean: 1
gray: mean: 25 sigma: 5
white: mean: 41 sigma: 4
Provide a raw estimation of means and standard deviations, then lock the file to prevent any further modification by the automatic procedure, using the validation procedure Validation Nobias Histo analysis or the command line :
"touch nobias_dionysos.han.loc"
We will try to simplify this interraction in a near future...
mri: Raw T1 MRI ( input )
brain_mask: T1 Brain Mask ( output )
variant: Choice ( input )
help: Choice ( input )
mri_corrected: T1 MRI Bias Corrected ( input )
histo_analysis: Histo Analysis ( input )
Commissure_coordinates: Commissure coordinates ( optional, input )
lesion_mask: 3D Volume ( optional, input )
first_slice: Integer ( input )
last_slice: Integer ( input )
Toolbox : Morphologist
User level : 0
Identifier :
AnaT1toBrainMaskCorrectionSupported file formats :
mri :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagebrain_mask :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagemri_corrected :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo AnalysisCommissure_coordinates :Commissure coordinates, Commissure coordinateslesion_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 image