


Compute a mesh of one or two hemispheres
This process create a mesh of each cerebral hemisphere. It include a pretreatment, which open the sulcus.
This new version yields a spherical mesh, which allows the inflation of the mesh as the grey/white mesh.
Side: Choice ( input )Left, Right or Both Hemispheres
mri_corrected: T1 MRI Bias Corrected ( input )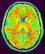
split_mask: Split Brain Mask ( input )
left_hemi_cortex: Left CSF+GREY Mask ( input )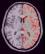
right_hemi_cortex: Right CSF+GREY Mask ( input )
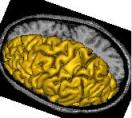
left_hemi_mesh: Left Hemisphere Mesh ( output )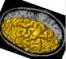
right_hemi_mesh: Right Hemisphere Mesh ( output )
fix_random_seed: Boolean ( input )
Toolbox : Morphologist
User level : 2
Identifier :
GetSphericalHemiSurfaceSupported file formats :
mri_corrected :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageleft_hemi_cortex :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageright_hemi_cortex :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageleft_hemi_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshright_hemi_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI mesh