A pipeline to process FSL connectome data of a subject into an individual connectivity matrix compatible with Constellation requirements.
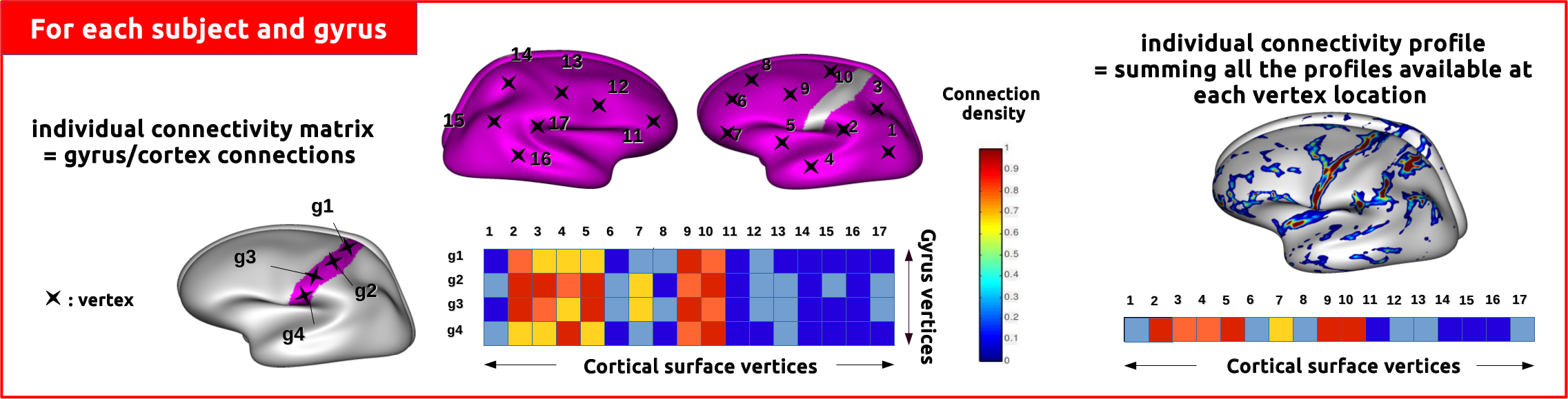
A complete pipeline: an intra_subject chain which builds the individual connectivity matrices. The preprocessings are done using FreeSurfer for the anatomical data, and FSL for the diffusion data.
The method implemented here is described in:
S. Lefranc, P. Roca, M. Perrot, C. Poupon, D. Le Bihan, J.-F. Mangin, and D. Rivière. Groupwise connectivity-based parcellation of the whole human cortical surface using watershed-driven dimension reduction. Medical Image Analysis, 30:11-29, 2016. [bibtex-entry]
Usage example
regions_nomenclature: /casa/build/share/brainvisa-share-4.6/nomenclature/translation/nomenclature_desikan_freesurfer.txtoutputs_database: /my/path/brainvisa_dbstudy_name: studyAmethod: averaged approachregion: lh.inferiorparietalprobtrackx_indir: /my/path/fsl_db/3T_fsl/omatrix1/100206temp_outdir: /tmpindividual_white_mesh: /my/path/freesurfer_db/100206/surf/bh.r.aims.white.giiregions_parcellation: /my/path/freesurfer_db/group_analysis/average_group/average_brain/bh.annot.averagebrain.giiregions_selection: Customkeep_regions: 'lh.unknown' 'lh.bankssts' 'lh.inferiorparietal'min_fibers_length: 20.0smoothing: 3.0kmax: 12normalize: Trueerase_matrix: True
regions_nomenclature: Nomenclature ROIs File ( input )Nomenclature of the cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas
outputs_database: OpenChoice ( input )This parameter retrieves all databases with brainvisa ontology present in your configuration, the generated files will be written on the chosen one (see the documentation to add a database in the BrainVisa configuration).
study_name: OpenChoice ( input )General name of the study.
method: Choice ( input )Two methods are proposed:
(1) averaged approach to obtain an average result on the group.
(2) concatenated approach to obtain individual results across the group.
region: OpenChoice ( input )The study region based on regions_nomenclature file.
probtrackx_indir: Directory ( input )Subjects directory in a FSL connectome database, where the fiber tracts files can be found.
temp_outdir: Directory ( input )Temporary directory where data files will be stored during importation phase.
Example: /tmp, but this directory size should be large enough to store all the temporary files during the process.
individual_white_mesh: White Mesh ( input )Freesurfer white-grey interface of the cortex.
Should not be inflated.
regions_parcellation: ROI Texture ( input )Cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas (?h.aparc.annot).
regions_selection: Choice ( input )This parameter can be used to exclude some regions from the data analysis. This is useful to study the specific connectivity between a given set of regions, or to exclude the initial patch region.
keep_regions: ListOf( OpenChoice ) ( input )Kept regions for the data analysis.
min_fibers_length: Float ( input )A filtering parameter to exclude low length fibers. Default to 20mm.
smoothing: Float ( input )Degree of smoothing (in millimetres).
Default to 3.0 mm.
kmax: Integer ( input )Maximal number of clusters used to parcellate the study region.
Default to 12.
normalize: Boolean ( input )By default the connectivity matrices values are normalized to balance any spurious weighting effects due to more connected regions or subjects, or to the tractography algorithm (number of seeds...). But sometimes we also like to see the raw connectivity matrix: in that case, uncheck this normalization.
erase_smoothed_matrix: Boolean ( input )
Toolbox : Constellation
User level : 1
Identifier :
constel_individual_pipeline_fsl_connectomeSupported file formats :
regions_nomenclature :Text file, Text fileprobtrackx_indir :Directory, Directorytemp_outdir :Directory, Directoryindividual_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshregions_parcellation :GIFTI file, GIFTI file, Texture