Create a 3D volume of the labelled sulci
This procedure creates 3D volumes including various parts of the sulci. For instance to compute SPAMs of their variability in the proportional system, or to use them as constraint in a warping process. In these volumes, each sulcus is represented by an integer label. These labels are associated either to the "name" attributes of the folds (manual or validated) or to the "label" attributes (automatic recognition). By default, the procedure uses "labels" if they exist, otherwise "name". This procedure is used with a cortical fold graph, whose nodes have been manually or automatically associated to a sulcus nomenclature.
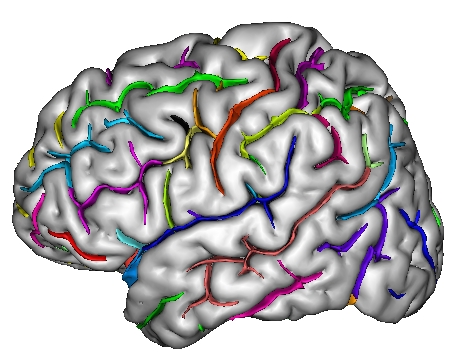

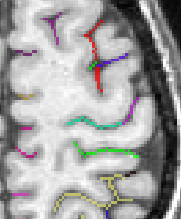
Each fold includes several buckets of voxels corresponding to the topologically simple surface making up its core (GREEN), its bottom lines (including branche's bottoms) (RED) and a few additional voxels corresponding to small or spurious branches (CYAN). Here is the example of a central sulcus:
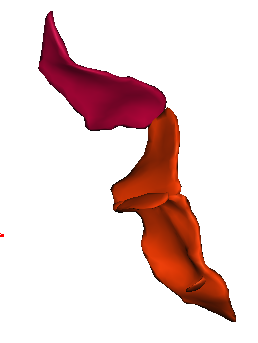
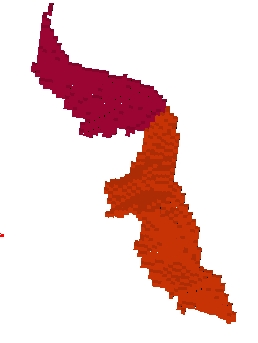
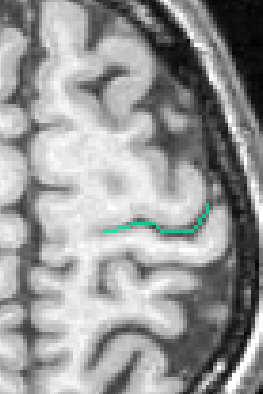
You can get images of the bottom, of the simple surface or the sum of everything. You can also get the junction between each sulcus and the brain hull, which corresponds to the line which is sometimes drawn manually by some teams. Anatomist's viewer show you the result (the eyes):
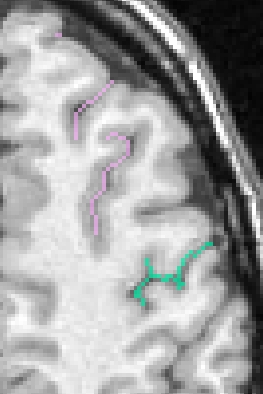
If you want to get only one sulcus, or a small list of sulci, write their label in the attibute "label_values". For instance "S.C._left" or "S.C._left S.F.sup._left":
graph: Cortical folds graph ( input )
mri: T1 MRI Bias Corrected ( optional, input )
transformation_matrix: Transformation matrix ( optional, input )
transformation_template: 3D Volume ( optional, input )
transformation: Choice ( optional, input )
binarize: Choice ( optional, input )
sulci: Sulci Volume ( output )
simple_surface: Simple Surface Volume ( optional, output )
bottom: Bottom Volume ( optional, output )
hull_junction: Hull Junction Volume ( optional, output )
compress: Choice ( input )
bucket: Choice ( input )
custom_buckets: String ( optional, input )
label_translation: Label translation ( optional, input )
input_int_to_label_translation: Log file ( optional, input )
int_to_label_translation: Log file ( optional, output )
label_attributes: Choice ( input )
custom_label_attributes: String ( optional, input )
node_edge_types: Choice ( input )
custom_node_edge_types: String ( optional, input )
label_values: String ( optional, input )
Toolbox : Morphologist
User level : 2
Identifier :
sulcuslabelvolumeFile name :
brainvisa/toolboxes/morphologist/processes/Sulci/Recognition/sulcuslabelvolume.pySupported file formats :
graph :Graph and data, Graph and datamri :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagetransformation_matrix :Transformation matrix, Transformation matrixtransformation_template :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesulci :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagesimple_surface :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagebottom :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagehull_junction :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagelabel_translation :Label Translation, Label Translationinput_int_to_label_translation :Text file, Text fileint_to_label_translation :Text file, Text file