In the NSA project, this process enables to individualize each brain in the matrix.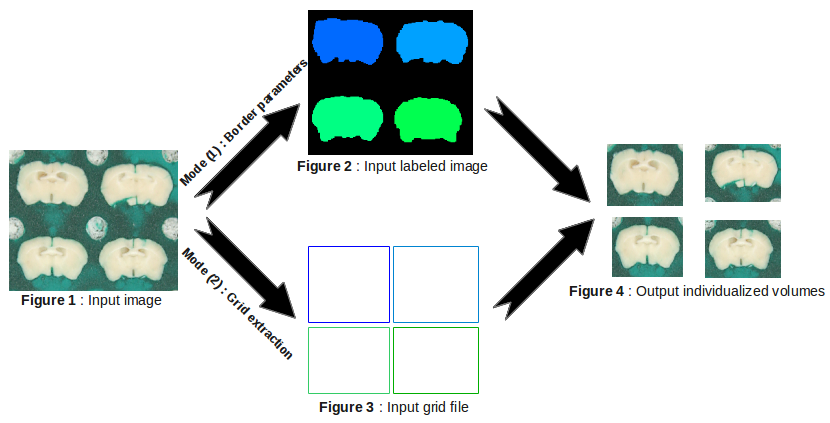
Input_ListOf_Image: ListOf( Volume 3D ) ( input )Upload list of 2d matrix(multiple brains) to be individualized.
Input_ListOf_BoundingBox: ListOf( Text file ) ( input )Select the list of the bounding box files.
Borders: ListOf( Réel ) ( optional, input )Enter the border size [sx sy sz] (default : [0,0,0] in mm).
Thickness: Réel ( input )
Output_Prefix: String ( optional, input )Enter the prefix that will be used to create the names of output volumes.
Output_Directory: Répertoire ( optional, entrée )Specify the output directory in which the volumes will be stored.
Output_ListOf_Volume: ListOf( Volume 3D ) ( output )Specify the list of output volumes.
Toolbox : Bioprocessing
Niveau d'utilisateur : 3
Identifiant :
IndividualizeLabeledImageNom de fichier :
brainvisa/toolboxes/bioprocessing/processes/research/toolbox/IndividualizeLabeledImage.pySupported file formats :
Input_ListOf_Image :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageInput_ListOf_BoundingBox :Minf, MinfOutput_Directory :Répertoire, RépertoireOutput_ListOf_Volume :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 image