
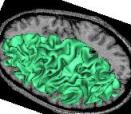
Control panel of the validation processes
T1mri: Raw T1 MRI ( input )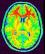
validation_total: Choice ( input )
mri_corrected: T1 MRI Bias Corrected ( input )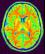
validation_mri_corrected: Choice ( input )
histo_analysis: Histo Analysis ( optional, input )
validation_histo_analysis: Choice ( input )
brain_mask: T1 Brain Mask ( optional, input )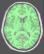
validation_brain_mask: Choice ( input )
split_mask: Split Brain Mask ( optional, input )
validation_split_mask: Choice ( input )
left_hemi_mesh: Left Hemisphere Mesh ( optional, input )
validation_left_hemi_mesh: Choice ( input )
right_hemi_mesh: Right Hemisphere Mesh ( optional, input )
validation_right_hemi_mesh: Choice ( input )
left_white_mesh: Left Hemisphere White Mesh ( optional, input )
validation_left_white_mesh: Choice ( input )
right_white_mesh: Right Hemisphere White Mesh ( optional, input )
validation_right_white_mesh: Choice ( input )
Toolbox : Morphologist
User level : 0
Identifier :
AnaValidateAllSupported file formats :
T1mri :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagemri_corrected :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysisbrain_mask :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_mask :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageleft_hemi_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshright_hemi_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshleft_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshright_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI mesh