A filtration of the study region fibers according to minimal and maximal allowed length, and a separation into two groups depending on whether one or two of their ends are connected to the mesh.
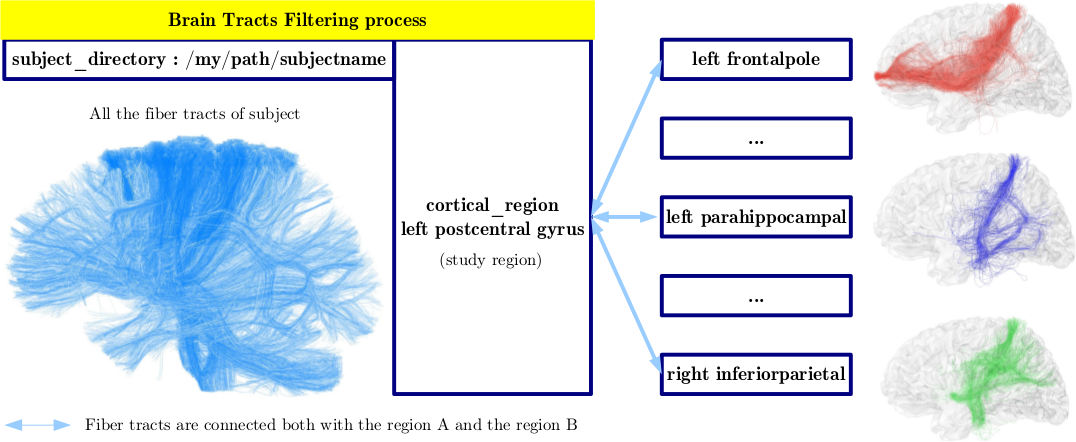
Depending on the starting region, the fiber tracts are filtered according to three criteria:
(1) only fibers connected to the starting region are retained (mandatory criteria)
(2) the fibers less than a threshold in length are removed (mandatory criteria)
(3) the fibers of longer than a threshold are removed (mandatory criteria)
This filtering is organized as follow:
- reads fibers: Only fibers generated by the Connectomist-2.0 and TrackVis softwares are accepted. It is possible to put a list of files.
- set gyrus pair name on each fiber: Each end of the fibers is associated with a vertex of the mesh, itself labeled by the starting region segmentation. This label is awarded to the fiber. If a fiber end is not identified on the mesh, it is labelled "notInMesh".
- select bundles from names: the fibers from at least one end to the starting region are kept.
- filter by length: smaller fibers than the threshold "minlength?" or larger than the threshold "maxlength_?" are deleted.
- merge the files: creating a single file output by fiber type (see below)
Two types of fiber tracts have to be taken into consideration in this process:
- labeled fibers are defined as having both ends attached to the mesh (and are consequently labelled)
- semilabeled fibers are defined as having only one end attached to the mesh (the other being not identified)
Usage example
regions_nomenclature: /casa/build/share/brainvisa-share-4.6/nomenclature/translation/nomenclature_desikan_freesurfer.txtoutputs_database: /my/path/brainvisa_dbstudy_name: studyAmethod: averaged approachregion: lh.inferiorparietalsubject_indir: /my/path/connectomist_db/B1500/StreamlineProbabilistic/aQBI/27seeds/001individual_white_mesh: /my/path/freesurfer_db/001/surf/bh.r.aims.white.giidw_to_t1: /my/path/connectomist_db/B1500/StreamlineProbabilistic/aQBI/27seeds/001/dw_to_t1.trmregions_parcellation: /my/path/freesurfer_db/group_analysis/average_group/average_brain/bh.annot.averagebrain.giifiber_tracts_format: bundlesmin_fibers_length: 20.0max_fibers_length: 500.0labeled_fibers: /my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/ default_tracking_session/connectivity_parcellation/avg/studyA/lh.inferiorparietal/filteredTracts/ 001_studyA_lh.inferiorparietal_labelled_fibers_20to500mm.bundlessemilabeled_fibers: /my/path/database_brainvisa/subjects/001/diffusion/default_acquisition/default_analysis/ default_tracking_session/connectivity_parcellation/avg/studyA/lh.inferiorparietal/filteredTracts/ 001_studyA_lh.inferiorparietal_semilabelled_fibers_20to500mm.bundles
regions_nomenclature: Nomenclature ROIs File ( input )Nomenclature of the cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas
outputs_database: Choice ( input )This parameter retrieves all databases with brainvisa ontology present in your configuration, the generated files will be written on the chosen one (see the documentation to add a database in the BrainVisa configuration).
study_name: OpenChoice ( input )General name of the study.
method: Choice ( input )Two methods are proposed:
(1) averaged approach to obtain an average result on the group.
(2) concatenated approach to obtain individual results across the group.
region: Choice ( input )The study region based on regions_nomenclature file.
subject_indir: Subject ( input )This is the subjects directory in a Connectomist database, where the fiber tracts files can be found.
individual_white_mesh: White Mesh ( input )Freesurfer white-grey interface of the cortex.
Should not be inflated.
dw_to_t1: Transform T2 Diffusion MR to Raw T1 MRI ( input )Affine spatial transformation to get the T1 MRI space from the dMRI diffusion (and tracts) space.
regions_parcellation: ROI Texture ( input )Cortical parcellation used to partition the study.
Example : Freesurfer Desikan_Killiany Atlas (?h.aparc.annot).
fiber_tracts_format: Choice ( input )Different fiber tracts formats are supported.
Selectable formats are:
(1) TrackVis file format with .trk as filename extension.
(2) Connectomist file format with .bundles as filename extension.
min_fibers_length: Float ( input )A filtering parameter to exclude low length fibers. Default to 20mm.
max_fibers_length: Float ( input )A filtering parameter to exclude long length fibers. Default to 500mm.
labeled_fibers: Filtered Fascicles Bundles ( output )All fibers having their ends identified on the mesh and corresponding to the various criteria in this process are merged into one output file.
semilabeled_fibers: Filtered Fascicles Bundles ( output )All fibers having only one end identified on the mesh and corresponding to the various criteria of this process are merged into one output file.
Toolbox : Constellation
User level : 2
Identifier :
constel_brain_tracts_filteringSupported file formats :
regions_nomenclature :Text file, Text filesubject_indir :Directory, Directoryindividual_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshdw_to_t1 :Transformation matrix, Transformation matrixregions_parcellation :GIFTI file, GIFTI file, Texturelabeled_fibers :Aims bundles, Aims bundlessemilabeled_fibers :Aims bundles, Aims bundles