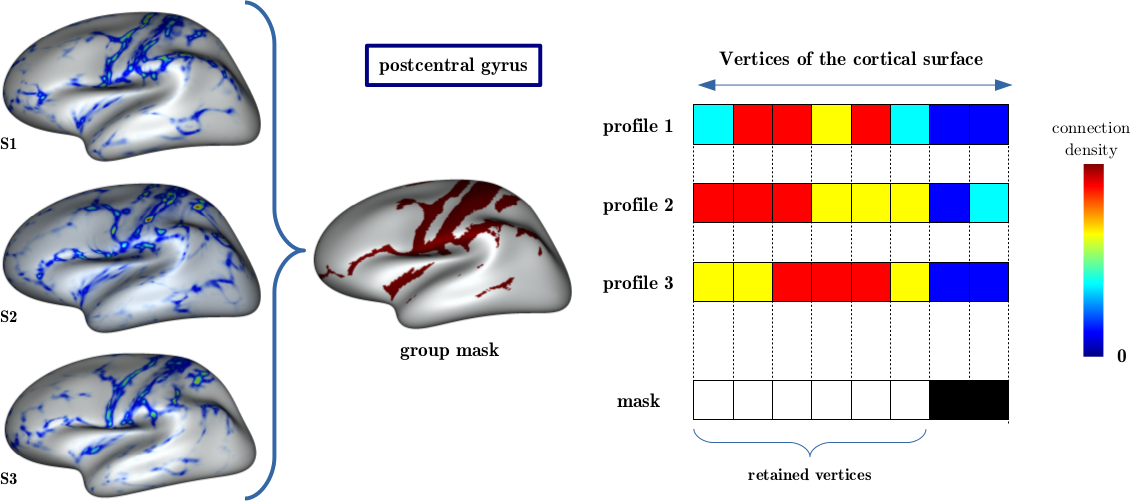
A mask is calculated to identify the cortical regions strongly connected and reproducible among subjects.
Usage example
new_study_name: group_studyAsubjects_group: /my/path/brainvisa_db/subjects/group_analysis/g1/g1_group.xmlmean_individual_profiles: '/my/path/brainvisa_db/subjects/001/diffusion/default_acquisition/default_analysis/default_tracking_session/connectivity_parcellation /avg/studyA/lh.inferiorparietal/clustering/smooth3.0/001_studyA_lh.inferiorparietal_mean_profile_20to500mm.gii'group_mask: /my/path/brainvisa_db/subjects/group_analysis/g1/connectivity_clustering/avg/studyA/lh.inferiorparietal/smooth3.0 /g1_avg_studyA_lh.inferiorparietal_mask_20to500mm.gii
new_study_name: String ( optional, input )Optional name given to the group study.
Default to study_name.
subjects_group: Group definition ( input )The XML file corresponding the subjects group.
mean_individual_profiles: ListOf( Connectivity Profile Texture ) ( input )List of the mean individual profiles of the given group of subjects.
group_mask: Mask Texture ( output )The most representative connectivity pattern of a group of subjects.
Toolbox : Constellation
User level : 2
Identifier :
constel_group_maskFile name :
brainvisa/toolboxes/constellation/processes/group_pipeline/tools/constel_group_mask.pySupported file formats :
subjects_group :XML, XMLmean_individual_profiles :GIFTI file, GIFTI file, Texturegroup_mask :GIFTI file, GIFTI file, Texture