This pipeline performs the registration of a template, and of its associated labels and probability maps, towards the target image.
Pipeline steps
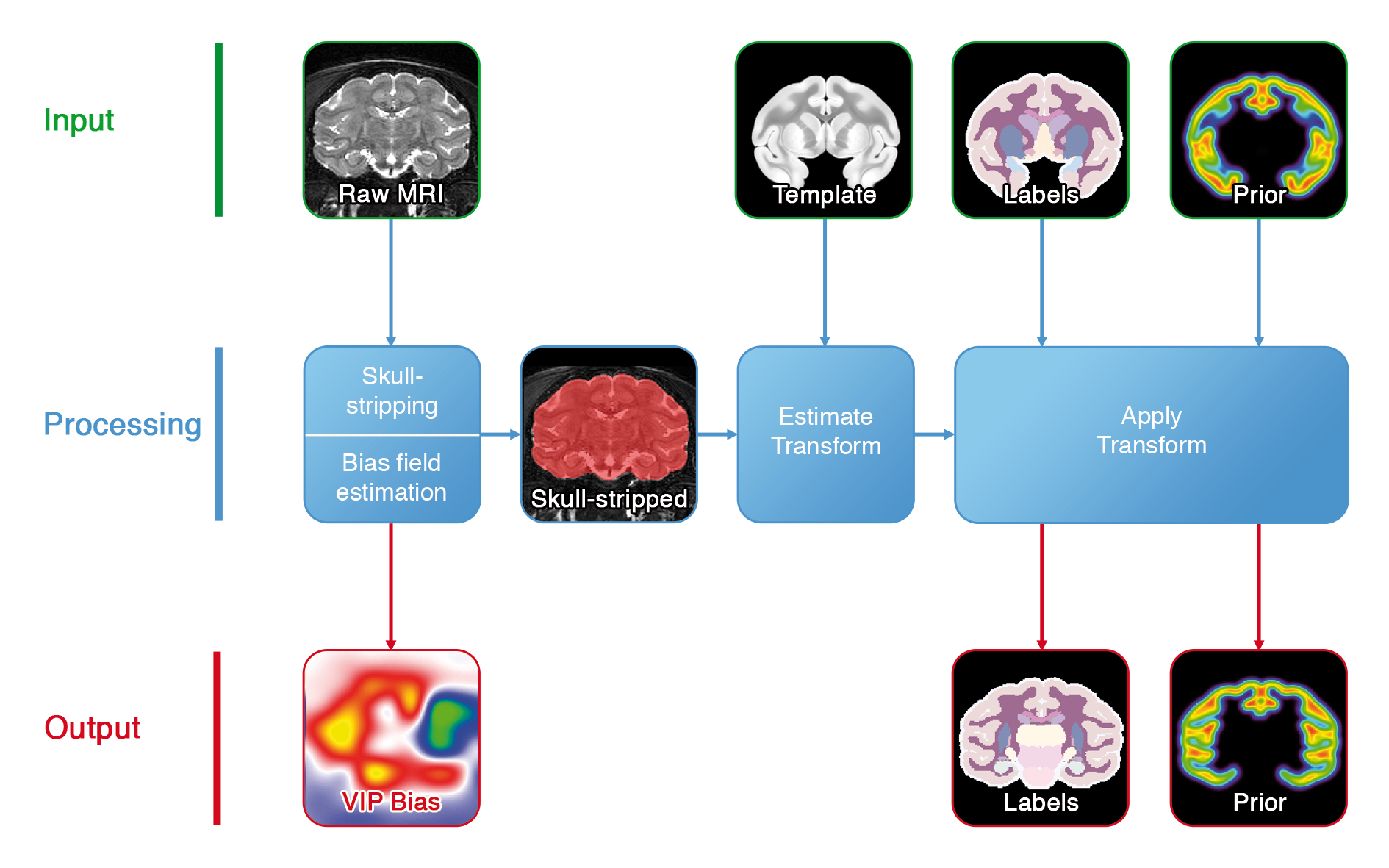
Figure 1. Registration pipeline.
- Prepare MRI: select anatomical landmarks to orient correctly the input volume.
- (Align & Resample): use landmarks to align the brain and resample it at a fixed resolution.
- VIP Bias Correction: non-statistical bias field estimation.
- Skull Stripping: compute a mask of the intracranial volume.
- Select Evaluation Points: select randomly a subset of points for the affine transform estimation.
- Atlas to MRI: Affine Transform Estimation
- Atlas to MRI: Non-Linear Transform Estimation
- Atlas to MRI: Apply Transforms
Landmarks selection
Before any computation is done, four anatomical landmarks should be manually selected:
Additional informations can be found in the MRI preparation documentation. This step is used to flip the MRI volume to BrainVISA's orientation.
- the anterior commissure ;
- the posterior commisure ;
- one point belonging to the interhemispheric plane ;
- one point belonging to the left hemisphere.
Optional alignment and resampling
Two optional steps, set in the Alignment & Resampling block, can be activated:
Additional information and rational for the activation or not of these options can be found in the Alignement & Resampling documentation.
- manually selected landmarks can be used to rigidly align the brain ;
- the MRI can be resampled so that it is segmented at a fixed resolution.
Registration
In order to make the process as robust as possible, several preprocessing steps are performed:
- the image is bias corrected with VIP's bias correction tool ;
- it is then skull-stripped with a combination of morphomathematical operations ;
- an affine transform is computed between the template and the skull-stripped image ;
- a non-linear transform is then computed between the template and the skull-stripped image ;
- the non-linear transformed is refined by registering the template towards the non skull-stripped image ;
- all atlas modalities are finally resampled in the target space ;
input_mri: P:MRI Raw ( input )Raw MRI
specie: OpenChoice ( optional, input )
contrast: OpenChoice ( optional, input )
is_mapping: Boolean ( optional, input )Set to true if the input image is a parametric map (T1 mapping, T2 mapping, etc) instead of a magnitude T1 or T2 weighted image.
point_anterior_commissure: Point3D ( input )Anterior commissure point. This should be AC's most posterior points that intersects the interhemispheric plane.
point_posterior_commissure: Point3D ( input )Posterior commissure point. This should be PC's most anterior points that intersects the interhemispheric plane.
point_interhemispheric: Point3D ( input )Any point from the interhemispheric plane located above (i.e. dorsal to) the AC-PC line.
point_left_hemisphere: Point3D ( optional, input )Any point located in the left hemisphere. If the MRI is in radiological orientation, this point should be located, on the screen, at the right of the interhemispheric plane.
template: P:Atlas Raw Template ( input )File containing the atlas template, i.e. the associated anatomical image (usually an averaged MR image).
labels: P:Atlas Raw Labels ( optional, input )File containing the atlas "hard" labels.
prior: P:Atlas Raw Prior ( optional, input )File containing the atlas probability maps.
lateralization: P:Atlas Raw Lateralization ( optional, input )File containing the atlas lateralization mask, i.e. a volumes of labels were voxels from the right hemisphere are labeled 1 and voxels from the left hemisphere are labeled 2.
align_and_resample: Boolean ( optional, input )If activated, AC-PC landmarks will be used to align the raw MRI. The raw MRI will also be resampled towards the atlas resolution.
resolution: Float ( optional, input )Output resolution of the Raw MRI. It will be resampled if different from the input resolution.
output_labels: P:Atlas Registered Labels ( optional, output )File containing the labels resampled in the target space.
output_prior: P:Atlas Registered Prior ( optional, output )File containing the probability maps resampled in the target space.
output_lateralization: P:Atlas Registered Lateralization ( optional, output )File containing the lateralization mask resampled in the target space.
Toolbox : Primatologist
User level : 0
Identifier :
primate_AtlasRegistrationPipelineFile name :
brainvisa/toolboxes/primatologist/processes/pipelines/primate_AtlasRegistrationPipeline.pySupported file formats :
input_mri :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagetemplate :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagelabels :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageprior :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagelateralization :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageoutput_labels :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imageoutput_prior :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imageoutput_lateralization :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 image