This pipeline helps you to choose the best reconstruction for a PET camera by quantifying activity in jaszczak's spheres.
Before using this pipeline:
All this steps can be openned separatly, using right click.
- Define your jaszczak if it is a new one.
- Import your CT and PET Dicom images
- Convert your dicom images into nii format.
Finally, you can use this pipeline by selecting just your CT image (others parameters are automatically deducted).
The Jaszczak SPECT Phantom provides consistent performance information for any SPECT or PET system.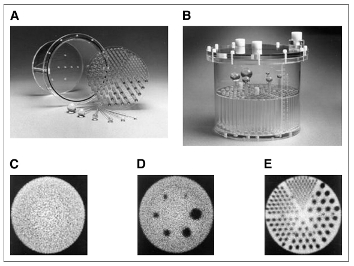
D : radioactivity is injected in spheres called "hot spheres", the others are "cold" ( you can see these ones in black). Then the activity in spheres' image is quantified and compare with the theoretical one, for each reconstruction. A graph allows you to compare reconstructions E : Rods are used to check visually the image resolution. To do so, all slices with rods are summed. To quantify spheres:
To quantify the activity in Jaszczak's spheres, you need to fill the fields about the activity injected in Jaszczak and its spheres.quantify spheres.
Pipeline steps' details :
The acquisition can be dynamic, so the mean is computed (step 1) All PET images and the CT one are segmented to check a possible gap between them. (step 2 Segment sph PET , 3 Segment sph CT, 4 Check fusion PET CT) Note : If so, the camera should be fixed. According to this gap, the spheres' mask is computed (step 5 Build Sph Mask) and used to quantify spheres (step 7 Quantify spheres) All slices with rods are summed and then displayed, to check visually the resolution (step 6 Sum rods) All reconstructions are quantified (step 7 Quantify spheres), using ROI previously computed and AimsRoiFeatures All reconstructions can be compared thanks to the graphic computed in step 8 Compare quantification
Pipeline steps' documentation :
Spheres Segmentations :
Display CT, PET, ROI and Mask fusions
Reconstruction Comparison :
database: Choice ( input )
display_separatly_each_process: Choice ( input )
Pets_mean: ListOf( Phantom PET Mean ) ( input )
CT: Phantom CT ( input )
phantom: String ( input )
center: String ( input )
jzkSpec: Phantom Spec ( input )
theoric_values: Phantom Theoric Values ( optional, output )
bkg_preparedActivity: Float ( input )prepared activity in background unit is Mega bequerel
bkg_H_M_pre: String ( input )
bkg_residualActivity: Float ( input )residual activity in background unit is Mega bequerel
bkg_H_M_res: String ( input )
sph_preparedActivity: Float ( input )prepared activity in spheres unit is Mega bequerel
sph_H_M_pre: String ( input )
sph_residualActivity: Float ( input )residual activity in spheres unit is Mega bequerel
sph_H_M_res: String ( input )
H_M_acq: String ( input )
periode_isotope: Float ( input )
run_parallel: Boolean ( input )
verbose: Boolean ( input )
reconstructionNames: ListOf( String ) ( input )
Pets_segmented: ListOf( Phantom PET Mean Segmented ) ( output )
ct_segmented: Phantom CT Segmented ( output )
sph_rois: Phantom ROI Sph ( optional, output )
sph_roi_in_Noise: Phantom ROI Sph inNoise ( output )
PET_mean_toDisplay: Phantom PET Mean ( output )
Pet_segmented_toDisplay: Phantom PET Mean Segmented ( output )
CT2PetMeans: ListOf( CT to PET transformation file ) ( output )
PETs_ghosts: ListOf( Phantom ROI Sph Ghosts ) ( output )
allRods: ListOf( Phantom Rods ) ( output )
allRods_sum: ListOf( Phantom Rods Sum ) ( output )
stats_inSpheres: ListOf( Phantom Activity Stats ) ( output )
stats_inNoise: ListOf( Phantom Noise Stats ) ( output )
stats_inGhosts: ListOf( Phantom ROI Sph Ghosts Stat ) ( output )
ratios_inSpheres: ListOf( Phantom Result ) ( output )
ratios_inGhosts: ListOf( Phantom Sph Ghosts Result ) ( output )
graph: Phantom Graph HotSph ( output )
graph_coldSph: Phantom Graph ColdSph ( output )
Spheres_ID_PVC: ListOf( Integer ) ( input )
Spheres_ID_FWHM: ListOf( Integer ) ( optional, input )
selected_FWHM_prieto: String ( optional, input )
selected_FWHM_normal: String ( optional, input )
selected_FWHM_special: String ( optional, input )
ROI_folders_list: ListOf( Phantom ROI PVC ) ( optional, output )
FWHM_informations_files_list: ListOf( FWHM Informations Jaszczak ) ( optional, output )
Chi_curves_folders_list: ListOf( Chi Square folder ) ( optional, output )
Corrected_uptakes_list: ListOf( Phantom PVC Corrected Uptakes Stats ) ( optional, output )
RC_calculation_method: Choice ( optional, input )
PVC_method: Choice ( optional, input )
Fill_centers: Choice ( optional, input )
CT_sph1_center: Point3D ( optional, input )
CT_sph2_center: Point3D ( optional, input )
CT_sph3_center: Point3D ( optional, input )
CT_sph4_center: Point3D ( optional, input )
CT_sph5_center: Point3D ( optional, input )
CT_sph6_center: Point3D ( optional, input )
PVC_dictionary: PVC Jaszczak dictionary ( optional, output )
Toolbox : Nuclear Imaging
User level : 0
Identifier :
JZKPipelineFile name :
brainvisa/toolboxes/nuclearimaging/processes/JZKPipeline.pySupported file formats :
Pets_mean :NIFTI-1 image, NIFTI-1 imageCT :NIFTI-1 image, NIFTI-1 imagejzkSpec :CSV file, CSV filetheoric_values :CSV file, CSV filePets_segmented :NIFTI-1 image, NIFTI-1 imagect_segmented :NIFTI-1 image, NIFTI-1 imagesph_rois :NIFTI-1 image, NIFTI-1 imagesph_roi_in_Noise :NIFTI-1 image, NIFTI-1 imagePET_mean_toDisplay :NIFTI-1 image, NIFTI-1 imagePet_segmented_toDisplay :NIFTI-1 image, NIFTI-1 imageCT2PetMeans :Transformation matrix, Transformation matrixPETs_ghosts :NIFTI-1 image, NIFTI-1 imageallRods :NIFTI-1 image, NIFTI-1 imageallRods_sum :NIFTI-1 image, NIFTI-1 imagestats_inSpheres :CSV file, CSV filestats_inNoise :CSV file, CSV filestats_inGhosts :CSV file, CSV fileratios_inSpheres :CSV file, CSV fileratios_inGhosts :CSV file, CSV filegraph :CSV file, CSV filegraph_coldSph :CSV file, CSV fileROI_folders_list :Directory, DirectoryFWHM_informations_files_list :Text file, Text fileChi_curves_folders_list :Directory, DirectoryCorrected_uptakes_list :CSV file, CSV filePVC_dictionary :Text file, Text file