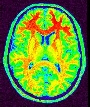



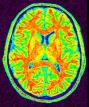
Correct for the spatial bias in usual MR images
This new bias correction is based on a method that should deal much better with low grey/white contrast and high bias magnitude.
t1mri: Raw T1 MRI ( input )
commissure_coordinates: Commissure coordinates ( optional, input )
sampling: Float ( input )
field_rigidity: Float ( input )weight for the field rigidity constraint
zdir_multiply_regul: Float ( input )
wridges_weight: Float ( input )
ngrid: Integer ( input )
background_threshold_auto: Choice ( input )
delete_last_n_slices: OpenChoice ( input )
t1mri_nobias: T1 MRI Bias Corrected ( output )
mode: Choice ( input )
write_field: Choice ( input )
field: T1 MRI Bias field ( optional, output )
write_hfiltered: Choice ( input )
hfiltered: T1 MRI Filtered For Histo ( output )
write_wridges: Choice ( input )
white_ridges: T1 MRI White Matter Ridges ( output )
variance_fraction: Integer ( input )
write_variance: Choice ( input )
variance: T1 MRI Variance ( output )
edge_mask: Choice ( input )
write_edges: Choice ( input )
edges: T1 MRI Edges ( output )
write_meancurvature: Choice ( input )
meancurvature: T1 MRI Mean Curvature ( optional, output )
fix_random_seed: Boolean ( input )
modality: Choice ( input )
use_existing_ridges: Boolean ( input )
Toolbox : Morphologist
User level : 0
Identifier :
T1BiasCorrectionFile name :
brainvisa/toolboxes/morphologist/processes/segmentationpipeline/components/T1BiasCorrection.pySupported file formats :
t1mri :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagecommissure_coordinates :Commissure coordinates, Commissure coordinatest1mri_nobias :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imagefield :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imagehfiltered :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imagewhite_ridges :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imagevariance :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imageedges :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imagemeancurvature :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 image