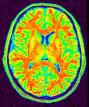

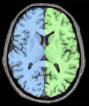



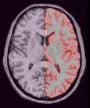
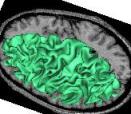
Compute a spherical mesh of the cortical surface
This treatment yields a spherical mesh of the cortical surface for each hemisphere. This mesh corresponds to the grey/white interface. This mesh can be inflated for visualization purpose: (cf Ana Inflate Cortical Surface).
The underlying algorithm relies on a series of morphological transformations, warping a parallepipedic box in which the hemisphere is embedded. (cf Ana Compute Cortical Fold Arg).
Example:
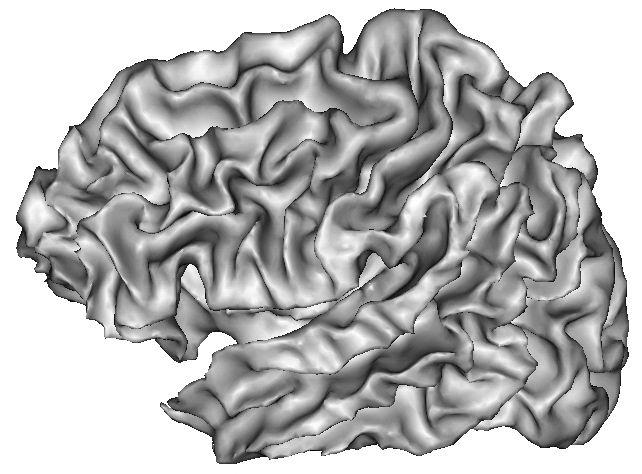
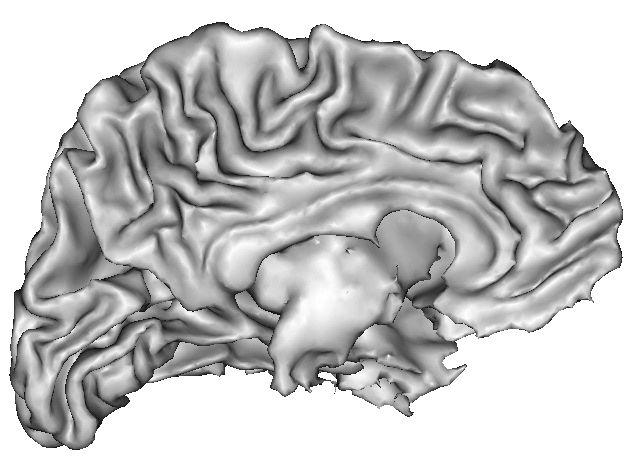
Because of spatial resolution, partial volume effect can lead to erosion of thin gyri, especially in frontal and temporal lobes. In such cases, you can improve the result via oversampling of the data, or increase pressure inside the surface. The risk is the creation of small objects growing out of the correct surface.
The parameters (iteration and ratio) control the smoothing of the mesh, which consists in moving each mesh node toward the barycenter of its neighbors (ratio tunes the amplitude of this motion). Too much smoothing may oversmooth thin gyri.
For more information :
From 3D magnetic resonance images to structural representations of the cortex topography using topology preserving deformations.
J.-F. Mangin, V. Frouin, I. Bloch, J. Regis, and J.Lopez-Krahe.
Journal of Mathematical Imaging and Vision, 5(4):297--318,1995.
Side: Choice ( input )
mri_corrected: T1 MRI Bias Corrected ( input )
histo_analysis: Histo Analysis ( input )
split_mask: Split Brain Mask ( input )
left_white_mesh: Left Hemisphere White Mesh ( output )
right_white_mesh: Right Hemisphere White Mesh ( output )
left_white_mesh_fine: Left Fine Hemisphere White Mesh ( output )
right_white_mesh_fine: Right Fine Hemisphere White Mesh ( output )
oversampling: Choice ( input )increase spatial resolution
pressure: Choice ( input )internal
iterations: Integer ( input )nb moves towards neighbor barycenter
rate: Float ( input )percentage of full move
Toolbox : Morphologist
User level : 2
Identifier :
AnaGetSphericalCorticalSurfaceSupported file formats :
mri_corrected :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysissplit_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageleft_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshright_white_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshleft_white_mesh_fine :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshright_white_mesh_fine :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI mesh