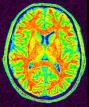

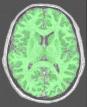




Split the brain into three parts (hemispheres + cerebellum)
This procedure aims at splitting the two hemispheres and at removing the cerebellum and a part of brain stem in order to give access to the internal and low faces of the cortex.
An erosion is applied to a mask of white matter in order to split it at the levels of corpus callosum and pons. This operation provides 3 seeds corresponding to the two hemispheres and cerebellum. Then, these seeds grow first inside white matter and finally throughout grey matter in order to recover the hemisphere shapes.
brain_mask: T1 Brain Mask ( input )
t1mri_nobias: T1 MRI Bias Corrected ( input )
histo_analysis: Histo Analysis ( input )
commissure_coordinates: Commissure coordinates ( optional, input )
use_ridges: Boolean ( input )
white_ridges: T1 MRI White Matter Ridges ( input )
use_template: Boolean ( input )
split_template: Hemispheres Template ( input )
mode: Choice ( input )
variant: Choice ( input )
bary_factor: Choice ( input )between 0 and 1.
mult_factor: Choice ( optional, input )
initial_erosion: Float ( input )
cc_min_size: Integer ( input )
split_brain: Split Brain Mask ( output )
fix_random_seed: Boolean ( input )
Toolbox : Morphologist
User level : 0
Identifier :
SplitBrainFile name :
brainvisa/toolboxes/morphologist/processes/segmentationpipeline/components/SplitBrain.pySupported file formats :
brain_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imaget1mri_nobias :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysiscommissure_coordinates :Commissure coordinates, Commissure coordinateswhite_ridges :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_template :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_brain :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 image