01. post mortem rebuilt 2D <=> 3D
Description
01. post mortem rebuilt 2D <=> 3D allows the 3D reconstruction of biological volumes arising from several histological and/or autoradiographic sections, beforehand individually digitized or acquired as overall scans (Cf. How to Scan).
Quality check
After running one of the processes of this category, you will be able to view the output files and assess the quality of the registrations and 3D reconstruction procedures in different orthogonal incidences by clicking on the  in Output_3dImage and visualizing volumes in Anatomist (Figures 2.1 and 2.2).
in Output_3dImage and visualizing volumes in Anatomist (Figures 2.1 and 2.2).
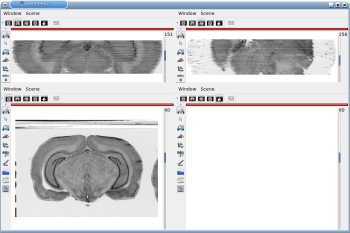
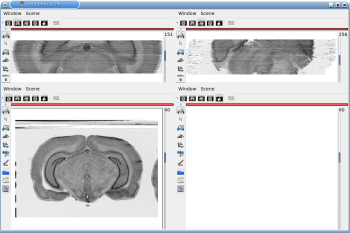
Fig. 2.1: 3D reconstruction of histological volume (Nissl stain) encompassing a large part of the visual system
(between bregma -5.2 mm and bregma -8.2 mm), in coronal, axial and sagittal views before (top) and after registration (bottom).
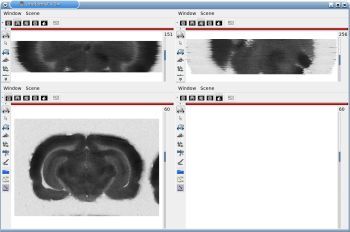
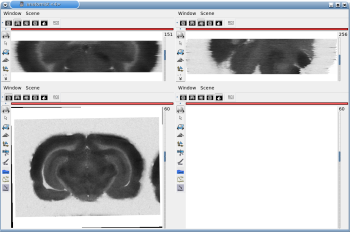
Fig. 2.2: 3D reconstruction of [14C]-2DG autoradiographic volume, encompassing a large part of the visual system (between bregma -5.2 mm and bregma -8.2 mm), in coronal, axial and sagittal views before (top) and after registration (bottom).
How To Scan : Optimized high throughput digitization procedure of autoradiographic and histological sections
Generality
Autoradiographs, corresponding histological section sets and radioactive [14C] standards are digitized with a flatbed scanner in transmission mode as either 8-bit or 16-bit grey-scale images (according to your scanner model).
In-plane digitization resolution needs to be sufficiently high to reveal the main structures of interest in the brain. Usually we choose a 600 dpi resolution (pixel size 42x42 mm2), for rat brain and a 1200 dpi resolution (pixel size 21x21 mm2), for the mouse one.
The calibration scale available with the scanner gives the relation between optical density and gray level values in the autoradiographic images. As gray level calibration is critical for quantitative autoradiography analysis, make sure to use appropriate software for acquiring your images.
Section arrangement on glass slides
A prerequisite for using BrainRAT process 02. image - individualizing and stacking multiple slices is that brain sections have been rigorously mounted on glass slides.
All the section arrangements on glass slides are conceivable (Figures 2.3 and 2.4).
Yet, the following condition shall be respected as far as possible:
ONLY ONE arrangement for the whole dataset
The section arrangements chosen by operator during the sectioning process will be requested as parameter values when using process 02. image - individualizing and stacking multiple slices.
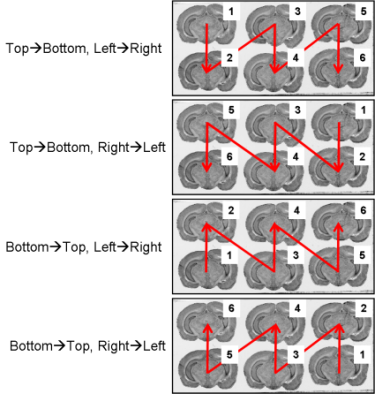
Fig. 2.3: All possible section arrangements on glass slides according to column first, which are managed by BrainRAT process 02. image - individualizing and stacking multiple slices.
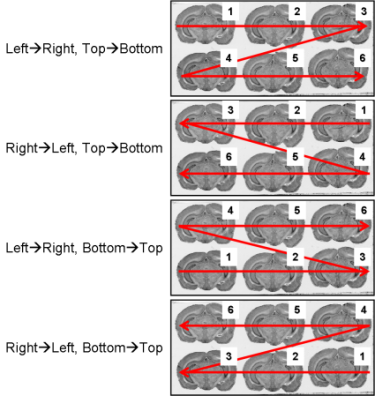
Fig. 2.4: All possible section arrangements on glass slides according to line first, which are managed by BrainRAT process 02. image - individualizing and stacking multiple slices.
The section arrangements chosen by operator during the sectioning process will be requested as parameter values when using process 02. image - individualizing and stacking multiple slices.
If for any reason you have not been able to respect this condition, an alternative is possible. Process each overall scan independently and then use process volume - subvolume stacking (03. toolbox) to concatenate the intermediate volumes (along Z-axis). Yet, make sure the volumes have all the same horizontal and vertical dimensions.
Number of sections per glass slide can vary: from 10-8 for small sections (olfactory bulb and frontal brain) to 6-4 for larger ones (midbrain and cerebellum). However, another condition shall be respected: acquiring and storing column images that have the same number of sections per glass slide.
NOTES:
- Though an automated analysis of width and height parameters for each section according to median values have been implemented to detect and to correct for vertical and horizontal overlaps between two sections, try to let enough space between each line and each column to limit such a phenomenom (Figure 2.5).
(a)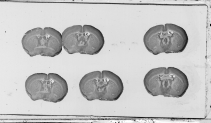 (b)
(b)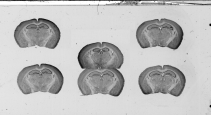
Fig. 2.5: Vertical (a) and horizontal (b) overlaps between two histological sections.
- High-quality sections are essential to make the corresponding volumes spatially consistent in 3D. Indeed, the block-matching registration technique is based on both the section edges and the whole image, so the result of the 3D reconstruction will strongly depend on the information available in the sections to be processed, that is to say, on the quality of data.
Glass slide arrangement on the surface of the scanner
Another prerequisite for using BrainRAT process 02. image - individualizing and stacking multiple slices is that the autoradiographic and histological sections have been acquired and stored under the form of glass slide column images called "overall scans" and whose size has been given by the scanner's field of view.
After the sectioning process, brain sections were exposed for several days to an autoradiographic film by arranging glass slides one below the other. Hence, the acquisition of autoradiographic sections can be achieved by digitizing the autoradiographic film, previously laid down on the surface of the scanner (Figure 2.6 (a)).
The acquisition of homologous histological sections (same sections as those processed for autoradiography or adjacent sections coming from another batch) can then be achieved by directly laying down glass slides, arranged one below the other, on the surface of the scanner (Figure 2.6 (b)).
(a) (b)
(b)
Fig. 2.6: (a) Acquisition of autoradiographic sections and corresponding radioactive [14C] standards. (b) Acquisition of homologous histological stained sections.
The arrangement in which the glass slides have been exposed to an autoradiographic film or laid down on the surface of the scanner (Top->Bottom or Bottom->Top) will be requested as a parameter value by process 02. image - individualizing and stacking multiple slices (Figure 2.7).
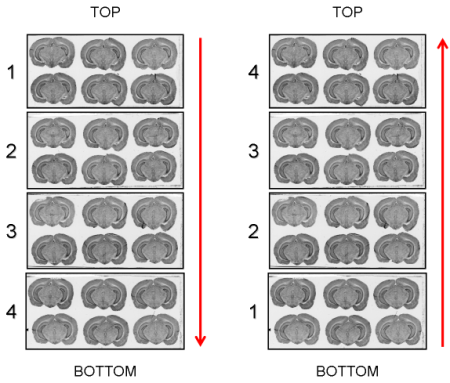
Fig. 2.7: Arrangement of glass slides on the surface of the scanner: Top->Bottom and Bottom->Top.
NOTES:
-
An a priori knowledge of section number and surface is used to perform iterative erosions, and thereby identify and extract the main connected components in the binary image of an overall scan. Consequently:
- There must not have objects with a similar surface in the image (e.g. handwritten section number);
- If important variations in section surface are observed from one glass slide to another, do not acquire too much glass slides together as a same overall scan;
- Changes in the original geometry of each section due to the mounting on glass slides may appear and induce the separation of some parts of the brain (olfactory bulb, occipital cortex, etc.). If necessary, a pre-processing can be required to artificially put back together these parts, using image manipulation software.
-
If several batches have been obtained from adjacent sections (several stain dyes, immunohistochemistry, etc.), make sure that the sections acquired together come from the same batch.
Each column of histological and autoradiographic sections as well as the standards are then independently digitized and stored in TIF format in a database (Figures 2.8 (a), (b) and (c)).
(a)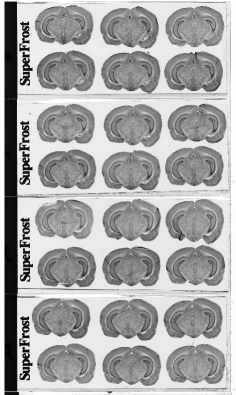 (b)
(b)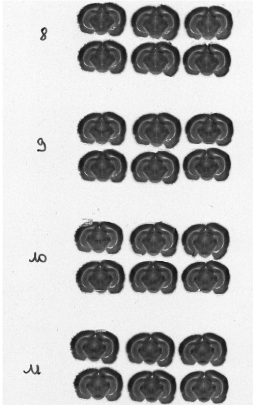 (c)
(c)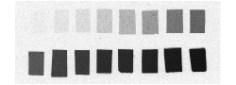
Fig. 2.8: (a) Overall scan of histological sections stained with cresyl violet (three glass slides arranged in a column, 600 dpi). (b) Overall scan of corresponding [14C]-2DG autoradiographic sections. (c) Radioactive [14C] standards..
Concerning our demonstration data set, we used a single 35*43 cm autoradiography film, the sections being distributed on the film as follows: 3 columns of 6 glass slides and 1 column of 7 glass slides, each one containing 6 sections, and 1 slide bearing radioactive [14C] standards within the fourth column. Hence, both [14C] standards and background were identical for all sections and consequently a single calibration was necessary.
![]() in Output_3dImage and visualizing volumes in Anatomist (Figures 2.1 and 2.2).
in Output_3dImage and visualizing volumes in Anatomist (Figures 2.1 and 2.2).





 (b)
(b)
 (b)
(b)

 (b)
(b) (c)
(c)