PyAnatomist examples¶
Most of these examples are expected to be run from an IPython shell, run from the directory of the examples (because some of them might load data from the current directory). Start IPython using:
>>> ipython -q4thread
Volume manipulation¶
Loading and viewing a Volume with anatomist
Download source: volumetest.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Volume manipulation
-------------------
Loading and viewing a Volume with anatomist
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.api as anatomist # works with all implementations
# initialize Anatomist
a = anatomist.Anatomist()
# load a volume in anatomist
avol = a.loadObject('irm.ima')
win = a.createWindow('Axial')
# put volume in window
a.addObjects([avol], [win])
# change palette
avol.setPalette('Blue-Red-fusion')
print("object and window are not deletable since there is a reference on it. Execute 'del win' and 'del avol' to delete them.")
# display in matplotlib for sphinx_gallery
win.sphinx_gallery_snapshot()
import sys
if 'sphinx_gallery' in sys.modules:
del win, avol
Aims / Anatomist volume manipulation¶
Loading, handling and viewing a Volume with aims and anatomist
Download source: aimsvolumetest.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
AIMS / Anatomist volume manipulation
====================================
Loading, handling and viewing a Volume with aims and anatomist
'''
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
from soma.qt_gui.qt_backend import Qt
import sys
# load any volume as a aims.Volume_* object
vol = aims.read('irm.ima')
runloop = False
if Qt.QApplication.instance() is None:
runloop = True
# initialize Anatomist
a = anatomist.Anatomist()
# convert the carto volume to Anatomist API
avol = a.toAObject(vol)
avol.releaseAppRef()
win = a.createWindow('Axial')
# put volume in window
win.addObjects(avol)
# volume change and update test
# using Numeric API
arr = vol.arraydata()
# we're just printing a white square in the middle of the volume
arr[0, 50:70, 100:130, 100:130] = 255
# update Anatomist object and its views
avol.setChanged()
avol.notifyObservers()
# fusion test
# have a second volume. We could have loaded another one
# (and it would have been nicer on screen). This example also
# shows the ability to share the volume data with multiple anatomist/aims
# objects
avol2 = a.toAObject(vol)
avol2.releaseAppRef()
# set a different palette on the second object so we can see something
a.setObjectPalette(avol2, 'Blue-Red-fusion')
# another view
win2 = a.createWindow('Axial')
a.addObjects(avol2, win2)
# create the fusion object
fus = a.fusionObjects([avol, avol2], 'Fusion2DMethod')
# show it
win3 = a.createWindow('Sagittal')
a.addObjects(fus, win3)
# display in matplotlib for sphinx_gallery
win3.sphinx_gallery_snapshot()
if runloop and 'sphinx_gallery' not in sys.modules:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del win, win2, win3, fus, avol, arr, avol2
Mesh manipulation¶
Using a mesh and modifying it while it is displayed
Download source: meshtest.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Mesh manipulation
-----------------
Using a mesh and modifying it while it is displayed
'''
from __future__ import absolute_import
from soma import aims
import time
import anatomist.direct.api as anatomist
import sys
from soma.qt_gui.qt_backend.QtGui import qApp
from soma.qt_gui.qt_backend import Qt
import six
# create a unit sphere of radius 1 and 500 vertices
m = aims.SurfaceGenerator.sphere(aims.Point3df(0, 0, 0), 1, 500, False)
# Multiply the sphere size by 100
for p in six.moves.xrange(m.vertex().size()):
m.vertex()[p] *= 100
runloop = Qt.QApplication.instance() is None and 'IPython' not in sys.modules
# Open Anatomist
a = anatomist.Anatomist()
# Put the mesh in anatomist
am = a.toAObject(m)
am.releaseAppRef()
# Create a new 3D window in Anatomist
aw = a.createWindow('3D')
# c = anatomist.CreateWindowCommand( '3D' )
# proc.execute( c )
# aw = c.createdWindow()
# Put the mesh in the created window
a.addObjects(am, aw)
# c = anatomist.AddObjectCommand( [ am ], [ aw ] )
# proc.execuFalsete( c )
# keep a copy of original vertices
coords = [aims.Point3df(m.vertex()[i])
for i in six.moves.xrange(len(m.vertex()))]
# take one vertex out of 3
points = six.moves.xrange(0, len(coords), 3)
for i in six.moves.xrange(10):
# shrink
for s in reversed(six.moves.xrange(100)):
for p in points:
m.vertex()[p] = coords[p] * s / 100.
am.setChanged()
am.notifyObservers()
qApp.processEvents()
time.sleep(0.01)
# expand
for s in six.moves.xrange(100):
for p in points:
m.vertex()[p] = coords[p] * s / 100.
am.setChanged()
am.notifyObservers()
qApp.processEvents()
time.sleep(0.01)
# display in matplotlib for sphinx_gallery
s = 0
for p in points:
m.vertex()[p] = coords[p] * s / 100.
am.setChanged()
am.notifyObservers()
if aw.sphinx_gallery_snapshot():
runloop = False
if runloop:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del aw, am, m
Anatomist commands¶
Using the commands interpreter in a generic way
Download source: customcommands.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Anatomist commands
------------------
Using the commands interpreter in a generic way
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
from soma.qt_gui.qt_backend import Qt
import sip
import sys
run_loop = Qt.QApplication.instance() is None \
and 'sphinx_gallery' not in sys.modules
a = anatomist.Anatomist()
cx = anatomist.cpp.CommandContext.defaultContext()
# custom command
i = cx.freeID()
c = a.execute('CreateWindow', type='Coronal', res_pointer=i)
win = c.createdWindow()
# otherwise, we can retreive the new window using the context
# (which will work if the exact command has no python binding)
print(cx.id(win))
print(cx.object(i))
win2 = cx.object(i)
print(win2 is win)
vol = aims.read('irm.ima')
avol = a.toAObject(vol)
avol.releaseAppRef()
# keywords / real objects test
a.execute('AddObject', objects=[avol], windows=[win])
if run_loop:
Qt.QApplication.instance().exec_
if run_loop or 'sphinx_gallery' not in sys.modules:
del win, win2, avol, vol, cx, i
Fusion 3D¶
Merging a volume and mesh in a 3D fusion object
Download source: fusion3D.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Fusion3D example
================
Fusion3D (mesh + texture from a volume) in Anatomist
'''
import anatomist.api as anatomist
# to expand the mesh bigger
from soma import aims
from soma.qt_gui.qt_backend import Qt
import numpy as np
import sys
runloop = False
if Qt.QApplication.instance() is None:
runloop = True
# initialize Anatomist
a = anatomist.Anatomist()
# load a volume in anatomist
avol = a.loadObject('irm.ima')
amesh = a.loadObject('test.mesh')
# this is to make the mesh bigger compared to the volume size
tr = aims.AffineTransformation3d()
tr.fromMatrix(np.matrix([[8., 0, 0, 204.4],
[0, 8., 0, 132.5],
[0, 0, 8., 68.4]]))
aims.SurfaceManip.meshTransform(amesh.toAimsObject(), tr)
amesh.UpdateMinAndMax()
# fusion the objects
fusion = a.fusionObjects(objects=[avol, amesh], method="Fusion3DMethod")
# params of the fusion
a.execute("Fusion3DParams", object=fusion,
method="line", submethod="mean", depth=4, step=1)
# open a window
win = a.createWindow('Axial')
# put volume in window
a.addObjects([fusion], [win])
# export the fusion texture in a file.
fusion.exportTexture("fusion.tex")
# display in matplotlib for sphinx_gallery
win.sphinx_gallery_snapshot()
if runloop and 'sphinx_gallery' not in sys.modules:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del fusion, win, amesh, avol, tr
Graph manipulation¶
Loading and displaying a graph
Download source: graph.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Graph manipulation
------------------
Loading and displaying a graph
'''
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
import sys
from soma.qt_gui import qt_backend
qt_backend.set_qt_backend(compatible_qt5=True)
from soma.qt_gui.qt_backend import QtGui
import sys
if QtGui.qApp.startingUp():
runqt = True
g = aims.read('Rbase.arg')
a = anatomist.Anatomist()
ag = a.toAObject(g)
ag.releaseAppRef()
for x in g.vertices():
x['toto'] = 12.3
g.vertices().list()[10]['toto'] = 24.3
g.vertices().list()[12]['toto'] = 48
ag.setColorMode(ag.PropertyMap)
ag.setColorProperty('toto')
ag.notifyObservers()
w = a.createWindow('3D')
w.addObjects(ag, add_graph_nodes=True)
w.camera(view_quaternion=[0.5, -0.5, -0.5, 0.5])
w.windowConfig(view_size=[642, 384])
if __name__ == '__main__':
# display in matplotlib for sphinx_gallery
if w.sphinx_gallery_snapshot():
runqt = False
if runqt:
QtGui.qApp.exec_()
if runqt or 'sphinx_gallery' in sys.modules:
del w, x, ag, g
Graph building¶
Creating a complete graph and nomenclature in Python
Download source: graph_building.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Graph building
--------------
Creating a complete graph and nomenclature in Python
'''
import anatomist.direct.api as ana
from soma import aims
from soma.qt_gui.qt_backend import Qt
import sys
# create graph structure
graph = aims.Graph('RoiArg')
# these are needed for internal conversions
graph['type.global.tri'] = 'roi.global.tri'
graph['roi.global.tri'] = 'roi roi_global.gii aims_mesh'
# create 2 nodes
v = graph.addVertex('roi')
mesh = aims.SurfaceGenerator.sphere([0, 0, 0], 10, 100)
aims.GraphManip.storeAims(graph, v, 'aims_mesh', mesh)
v['name'] = 'sphere1'
v = graph.addVertex('roi')
mesh = aims.SurfaceGenerator.sphere([0, 0, 100], 10, 100)
aims.GraphManip.storeAims(graph, v, 'aims_mesh', mesh)
v['name'] = 'sphere2'
# create a corresponding nomenclature
hie = aims.Hierarchy()
hie.setSyntax('hierarchy')
hie['graph_syntax'] = 'RoiArg'
n = aims.Tree(True, 'fold_name')
n['name'] = 'sphere1'
n['color'] = aims.vector_S32([255, 255, 0])
hie.insert(n)
n = aims.Tree(True, 'fold_name')
n['name'] = 'sphere2'
n['color'] = aims.vector_S32([0, 255, 0])
hie.insert(n)
runloop = Qt.QApplication.instance() is not None
# create anatomist objects
a = ana.Anatomist()
ahie = a.toAObject(hie)
ahie.releaseAppRef()
agraph = a.toAObject(graph)
agraph.releaseAppRef()
# display graph
w = a.createWindow('3D')
w.addObjects(agraph, add_children=True)
br = a.createWindow('Browser')
br.addObjects(ahie)
w.sphinx_gallery_snapshot()
if runloop and 'sphinx_gallery' not in sys.modules:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del w, br, agraph, ahie, v, graph, hie, n
Events handling¶
Catching click events and plugging a callback
Download source: events.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Events handling
---------------
Catching click events and plugging a callback
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
a = anatomist.Anatomist()
# definig a custom event handler in python:
def clickHandler(eventName, params):
print('click event: ', eventName)
print('LinkedCursor event contents:', list(params.keys()))
pos = params['position']
print('pos:', pos)
win = params['window']
print('window:', win)
# register the function on the cursor notifier of anatomist. It will be
# called when the user click on a window
a.onCursorNotifier.add(clickHandler)
# definig a custom event in python
class TotoEvent (anatomist.cpp.OutputEvent):
def __init__(self):
# we can't make a custom Object yet...
anatomist.cpp.OutputEvent.__init__(self, 'Toto',
{}, 1)
ev = TotoEvent()
ev.send()
# ...
# wen you're done
# you can remove the handler
# a.onCursorNotifier.remove(clickHandler)
Selection handling¶
Getting selected objects
Download source: selection.py
# -*- coding: utf-8 -*-
'''
Selection handling
------------------
Getting selected objects
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as ana
from soma import aims
from soma.aims import colormaphints
import sys
import os
# determine wheter we are using Qt4 or Qt5, and hack a little bit accordingly
from soma.qt_gui import qt_backend
from soma.qt_gui.qt_backend import QtCore, QtGui
# do we have to run QApplication ?
if not QtGui.QApplication.instance() and 'sphinx_gallery' not in sys.modules:
runqt = True
else:
runqt = False
# start the Anatomist engine, in batch mode (no main window)
a = ana.Anatomist('-b')
# create a sphere mesh
m = aims.SurfaceGenerator.sphere(aims.Point3df(0), 100, 100)
mesh = a.toAObject(m)
mesh.releaseAppRef()
# Create a new 3D window in Anatomist
aw = a.createWindow('3D')
# Put the mesh in the created window
a.addObjects(mesh, aw)
g = a.getDefaultWindowsGroup()
print('mesh isSelected:', g.isSelected(mesh))
print('selecting it')
g.setSelection(mesh)
print("selection in default group", a.getSelection())
print("selection of", g, g.getSelection())
sel = g.getSelection()
# print(mesh, sel, mesh == sel[0], mesh is sel[0])
# print('mesh isSelected:', g.isSelected( mesh ))
# run Qt
if runqt:
qapp.exec_()
elif 'sphinx_gallery' in sys.modules:
aw.sphinx_gallery_snapshot()
if runqt or 'sphinx_gallery' in sys.modules:
del aw, mesh, m, g, sel
Selection by nomenclature¶
Selecting graph nodes according a nomenclature
Download source: nomenclatureselection.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Selection by nomenclature
-------------------------
Selecting graph nodes according a nomenclature
'''
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
from soma.qt_gui.qt_backend import Qt
import os
import sys
runloop = Qt.QApplication.instance() is None
a = anatomist.Anatomist()
sh = aims.carto.Paths.shfjShared()
nom = a.loadObject(os.path.join(sh, 'nomenclature', 'hierarchy',
'sulcal_root_colors.hie'))
graph = a.loadObject('Rbase.arg')
w = a.createWindow('3D')
w.addObjects(graph, add_graph_nodes=True)
a.execute('SelectByNomenclature', names='PREFRONTAL_right', nomenclature=nom)
# to unselect all
# a.execute( 'Select' )
if 'sphinx_gallery' in sys.modules:
# display in matplotlib for sphinx_gallery
w.camera(view_quaternion=[0.5, -0.5, -0.5, 0.5])
w.windowConfig(view_size=[642, 384])
w.sphinx_gallery_snapshot()
runloop = False
if runloop:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del w, graph, nom
Sphere example¶
Subclassing Anatomist objects
Download source: sphere.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Sphere example
--------------
Subclassing Anatomist objects
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
import six
class ASphere(anatomist.cpp.ASurface_3):
def __init__(self, mesh=None):
self._center = aims.Point3df(0, 0, 0)
self._radius = 100
if mesh is not None:
if type(mesh) is six.string_types:
# mesh is a filename: read it
anatomist.cpp.ASurface_3.__init__(self, mesh)
r = aims.Reader()
m = aims.rc_ptr_AimsTimeSurface_3(r.read(mesh))
self.setSurface(m)
else:
# mesh should be an Aims mesh: assign it
anatomist.cpp.ASurface_3.__init__(self)
self.setSurface(mesh)
else:
# generate a sphere mesh
anatomist.cpp.ASurface_3.__init__(self)
m = aims.rc_ptr_AimsTimeSurface_3(aims.SurfaceGenerator.sphere(
self._center, self._radius, 100))
self.setSurface(m)
def radius(self):
return self._radius
def center(self):
return self._center
def setCenter(self, c):
if type(c) is not aims.Point3df:
c = aims.Point3df(*c)
cdiff = c - self._center
self._center = c
# translate all
v = self.surface().vertex()
for vi in v:
vi += cdiff
self.setChanged()
self.UpdateMinAndMax()
self.notifyObservers()
def setRadius(self, r):
if r == 0:
print('can\'t assign radius 0')
return
scl = float(r) / self._radius
self._radius = float(r)
# scale all
v = self.surface().vertex()
c = self._center
for vi in v:
vi.assign((vi - c) * scl + c)
self.setChanged()
self.UpdateMinAndMax()
self.notifyObservers()
# example
if __name__ == '__main__':
a = anatomist.Anatomist()
s = ASphere()
s.setName('sphere')
a.registerObject(s)
# import qt
# qt.qApp.exec_loop()
Ellipsoid example¶
Updating the size and shape of an object interactively
Download source: ellipsoid.py
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Ellipsoid example
-----------------
Updating the size and shape of an object interactively
'''
from __future__ import absolute_import
from soma import aims
import anatomist.direct.api as anatomist
import time
import numpy
import sys
from six.moves import range
from soma.qt_gui import qt_backend
from soma.qt_gui.qt_backend import QtGui, Qt
if 'IPython' in sys.modules and Qt.QApplication.instance() is None:
run_loop = False
else:
run_loop = True
a = anatomist.Anatomist()
def rotation_3d(a1, a2, a3):
'''
Return matrix 4x4 of rotation along 3 canonic axis based on 3 angles.
- a1, a2, a3 : rotation angles.
'''
c1, s1 = numpy.cos(a1), numpy.sin(a1)
c2, s2 = numpy.cos(a2), numpy.sin(a2)
c3, s3 = numpy.cos(a3), numpy.sin(a3)
m1 = numpy.matrix([
[c1, -s1, 0, 0],
[s1, c1, 0, 0],
[0, 0, 1, 0],
[0, 0, 0, 1]])
m2 = numpy.matrix([
[1, 0, 0, 0],
[0, c2, -s2, 0],
[0, s2, c2, 0],
[0, 0, 0, 1]])
m3 = numpy.matrix([
[c3, 0, -s3, 0],
[0, 1, 0, 0],
[s3, 0, c3, 0],
[0, 0, 0, 1]])
return m1 * m2 * m3
def apply_transform(mesh, scaling, rot, translate):
'''
Transform mesh with scaling, translation and rotation matrix.
'''
mesh2 = aims.AimsTimeSurface_3(mesh)
# create transformation
scale = numpy.asmatrix(numpy.diag(scaling))
rot[:, 3] = numpy.asmatrix(translate).T
cov = rot * scale * rot.I
# apply transformation
motion = aims.Motion(numpy.asarray(cov).flatten())
aims.SurfaceManip.meshTransform(mesh2, motion)
# update mesh in anatomist object
asphere.setSurface(mesh2)
def transform(mesh, angles, scaling, translate):
'''
Create and apply transformation to mesh.
'''
angles[0] += 0.02
angles[1] += 0.03
angles[2] += 0.05
a1, a2, a3 = angles
scaling = numpy.array([1.1 + numpy.cos(a1 * 3), 1.1 + numpy.cos(a2 * 2),
1.1 + numpy.cos(a3), 0]) * 0.5
scaling[3] = 1.
rot = rotation_3d(a1, a2, a3)
apply_transform(mesh, scaling, rot, translate)
mesh = aims.SurfaceGenerator.sphere(aims.Point3df(0, 0, 0), 1, 500)
asphere = a.toAObject(mesh)
asphere.releaseAppRef()
aw = a.createWindow('3D')
aw.setHasCursor(0)
a.addObjects([asphere], [aw])
if __name__ == '__main__':
angles = numpy.array([0., 0., 0.])
scaling = numpy.array([1., 1., 1., 0.])
translate = numpy.array([0, 0, 0, 1])
if 'sphinx_gallery' in sys.modules:
for i in range(90):
transform(mesh, angles, scaling, translate)
asphere.setChanged()
asphere.notifyObservers()
QtGui.qApp.processEvents()
# display in matplotlib for sphinx_gallery
import matplotlib
matplotlib.use('agg', force=True) # force agg
aw.imshow(show=True)
elif run_loop:
while a.getControlWindow().isVisible():
transform(mesh, angles, scaling, translate)
asphere.setChanged()
asphere.notifyObservers()
QtGui.qApp.processEvents()
time.sleep(0.01)
del aw, asphere, mesh
Custom controls example¶
Plugging new conrols / actions in Anatomist views
See also: Anatomist View / Controler model
Download source: control.py
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
# -*- coding: utf-8 -*-
'''
Custom controls example
-----------------------
Plugging new conrols / actions in Anatomist views
'''
import anatomist.direct.api as anatomist
from soma import aims
from soma.aims import colormaphints
import sys
import os
import math
sys.path.insert(0, '.')
# use the right version of Qt and of its python bindings (PyQt4/5/PySide)
from soma.qt_gui.qt_backend import QtCore, QtGui, loadUi
# do we have to run QApplication ?
if QtGui.qApp.startingUp() and 'IPython' not in sys.modules:
runqt = True
else:
runqt = False
import sphere
selmesh = None
selanamesh = None
class MyAction(anatomist.cpp.Action):
def name(self):
return 'MyAction'
def resetRadius(self):
print('reset radius to 1')
s.setRadius(1.)
def startMoveRadius(self, x, y, globx, globy):
print('start move radius', x, y)
self._initial = (x, y)
self._radius = s.radius()
def endMoveRadius(self, x, y, globx, globy):
print('end move radius', x, y)
def moveRadius(self, x, y, globx, globy):
print('move radius', x, y)
s.setRadius(math.exp(0.01 * (self._initial[1] - y)) * self._radius)
def takePolygon(self, x, y, globx, globy):
# print('takePolygon', x, y)
print('coucou', x, y)
w = self.view().aWindow()
obj = w.objectAtCursorPosition(x, y)
# print('object:', obj)
if obj is not None:
print('object:', obj, obj.name())
poly = w.polygonAtCursorPosition(x, y, obj)
# print('polygon:', poly)
mesh = anatomist.cpp.AObjectConverter.aims(obj)
# print('mesh:', mesh)
ppoly = mesh.polygon()[poly]
vert = mesh.vertex()
# print(ppoly[0], ppoly[1], ppoly[2])
# print(vert[ppoly[0]], vert[ppoly[1]], vert[ppoly[2]])
global selmesh, selanamesh
if selmesh is None:
selmesh = aims.AimsSurfaceTriangle()
selmesh.vertex().assign(
[vert[ppoly[0]], vert[ppoly[1]], vert[ppoly[2]]])
selmesh.polygon().assign([aims.AimsVector_U32_3(0, 1, 2)])
if selanamesh is None:
selanamesh = anatomist.cpp.AObjectConverter.anatomist(selmesh)
a = anatomist.Anatomist()
a.execute('SetMaterial', objects=[
selanamesh], diffuse=[0, 0, 1., 1.])
a.execute('AddObject', objects=[selanamesh], windows=[w])
selanamesh.setChanged()
selanamesh.notifyObservers()
class MyControl(anatomist.cpp.Control):
def __init__(self, prio=25):
anatomist.cpp.Control.__init__(self, prio, 'MyControl')
def eventAutoSubscription(self, pool):
key = QtCore.Qt
NoModifier = key.NoModifier
ShiftModifier = key.ShiftModifier
ControlModifier = key.ControlModifier
AltModifier = key.AltModifier
self.keyPressEventSubscribe(key.Key_C, NoModifier,
pool.action('MyAction').resetRadius)
self.mouseLongEventSubscribe(key.LeftButton, NoModifier,
pool.action('MyAction').startMoveRadius,
pool.action('MyAction').moveRadius,
pool.action('MyAction').endMoveRadius,
False)
self.mousePressButtonEventSubscribe(key.RightButton, NoModifier,
pool.action('MyAction').takePolygon)
a = anatomist.Anatomist()
qapp = QtGui.QApplication.instance()
pix = QtGui.QPixmap('control.xpm')
anatomist.cpp.IconDictionary.instance().addIcon('MyControl', pix)
ad = anatomist.cpp.ActionDictionary.instance()
ad.addAction('MyAction', lambda: MyAction())
cd = anatomist.cpp.ControlDictionary.instance()
cd.addControl('MyControl', lambda: MyControl(), 25)
cm = anatomist.cpp.ControlManager.instance()
cm.addControl('QAGLWidget3D', '', 'MyControl')
s = sphere.ASphere()
a.registerObject(s)
aw = a.createWindow('3D')
a.addObjects([s], [aw])
if 'sphinx_gallery' in sys.modules:
# display in matplotlib for sphinx_gallery
import matplotlib
matplotlib.use('agg', force=True) # force agg
aw.imshow(show=True)
runqt = False
# run Qt
if runqt:
qapp.exec_()
if runqt or 'sphinx_gallery' in sys.modules:
del aw, selanamesh, selmesh, s, ad, cd, cm, pix
Custom fusion example¶
Registering new fusion types
Download source: fusion.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Custom fusion example
=====================
Creating a new custom fusion type in Anatomist
'''
from soma import aims
import os
import anatomist.direct.api as anatomist
a = anatomist.Anatomist()
class MyAObjectKrakboumCallback(anatomist.cpp.ObjectMenuCallback):
def __init__(self):
anatomist.cpp.ObjectMenuCallback.__init__(self)
def doit(self, objects):
print('MyAObjectKrakboumCallback:', objects)
class MyAObject(anatomist.cpp.AObject):
_type = anatomist.cpp.AObject.registerObjectType('MyAObject')
_menus = None
icon = os.path.join(str(a.anatomistSharedPath()),
'icons', 'list_cutmesh.xpm')
ot = anatomist.cpp.QObjectTree
ot.setObjectTypeName(_type, 'Example of a custom AObject')
ot.setObjectTypeIcon(_type, icon)
def __init__(self, filename=''):
anatomist.cpp.AObject.__init__(self, filename)
self.setType(MyAObject._type)
self.setReferential(a.centralReferential())
def optionTree(self):
# set new option menus on this new object type
if MyAObject._menus is None:
m = anatomist.cpp.ObjectMenu()
krak = MyAObjectKrakboumCallback()
m.insertItem(['File'], 'Reload',
anatomist.cpp.ObjectActions.fileReloadMenuCallback())
m.insertItem(['Color'], 'Material',
anatomist.cpp.ObjectActions.colorMaterialMenuCallback())
m.insertItem(['Rototo', 'pouet'], 'krakboum', krak)
MyAObject._menus = m.releaseTree()
# avoid deleting the python part of the callback
MyAObject._nodelete = [krak]
return MyAObject._menus
class MyFusion(anatomist.cpp.FusionMethod):
def __init__(self):
anatomist.cpp.FusionMethod.__init__(self)
print("init myfusion")
def canFusion(self, objects):
print("MyFusion : canFusion")
return 120
def fusion(self, objects):
print("MyFusion : fusion")
return MyAObject()
def ID(self):
return 'myFusion'
def orderingMatters(self):
return False
# Register MyFusion
anatomist.cpp.FusionFactory.registerMethod(MyFusion())
# Load an object
obj = a.loadObject('test.mesh')
# if __name__ == '__main__' :
# import qt
# if qt.QApplication.startingUp():
# qt.qApp.exec_loop()
Anatomist API tests¶
Most of anatomist API module features
Download source: anatomistapiTests.py
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
"""
Anatomist api tests
===================
This example uses data of i2bm platform.
"""
from __future__ import print_function
from __future__ import absolute_import
import os
import sys
import soma.config
import numpy as np
import tempfile
data_directory = os.path.join(
os.environ.get("BRAINVISA_TEST_RUN_DATA_DIR", ""),
"tmp_tests_brainvisa/data_for_anatomist")
failures = []
mode = 'direct mode'
def testDirectImpl(interactive=True):
import anatomist.direct.api as pyanatomist
global mode
mode = 'direct mode'
a = pyanatomist.Anatomist()
return testAnatomist(a, interactive)
def testSocketImpl(interactive=True):
# test socket impl, not threaded
import anatomist.socket.api as pyanatomist
global mode
mode = 'socket mode'
a = pyanatomist.Anatomist()
return testAnatomist(a, interactive)
def test_assert(condition, message):
try:
assert(condition)
except AssertionError:
global failures
failures.append('%s: %s' % (mode, message))
print('** error: %s: %s **' % (mode, message))
def testAnatomist(a, interactive=True):
print("\n--- CreateWindowsBlock ---")
block = a.createWindowsBlock(3) # a block of windows with 3 columns
# in direct api the block object is really created only when the first
# window is added to it
print("block.internalRep = ", block.internalRep, ", block.nbCols = ",
block.nbCols)
test_assert(block.internalRep not in (None, 0), "block is invalid")
print("\n--- CreateWindow ---")
w1 = a.createWindow(wintype='Axial', block=block)
print("w1 : Axial window, added to the block. w1 = ", w1,
", block.internalRep = ", block.internalRep)
test_assert(w1 is not None, 'window w1 is None')
test_assert(w1.block == block, 'block in w1 is wrong')
w2 = a.createWindow(wintype='Sagittal', geometry=[10, 20, 200, 500],
block=block)
test_assert(w2 is not None, 'window w2 is None')
print(
"w2 : Sagittal window, added to the block. The geometry attribute is not taken into account because of the block, the window is resized to fit into the block.")
w3 = a.createWindow(wintype='Coronal', block=block, no_decoration=True)
print(
"w3 : Coronal window, added to the block (3rd column), thanks to the attribute no decoration, the window has no menus, buttons and so on...")
test_assert(w3 is not None, 'window w3 is None')
w4 = a.createWindow(wintype='3D', geometry=[10, 20, 200, 500])
print("w4 : 3D window, not in the block, with geometry attribute.")
test_assert(w4 is not None, 'window w4 is None')
w5 = a.createWindow(wintype='3D', no_decoration=True)
print("w5 : 3D window, not in the block, with no decoration.")
test_assert(w5 is not None, 'window w5 is None')
print("\n--- LoadObject ---")
o = a.loadObject(os.path.join(data_directory,
"subject01/subject01_Lwhite.mesh"), "objetO")
test_assert(o is not None, 'object o is None')
print("o : mesh Lhemi.mesh, renamed objecO, internalRep = ", o,
", o.__class__ = ", o.__class__)
o2 = a.loadObject(os.path.join(data_directory,
"subject01/subject01_Lwhite_curv.tex"),
"objetO2")
test_assert(o2 is not None, 'object o2 is None')
print("o2 : texture Lwhite_curv.tex, renamed objetO2, internalRep = ", o2)
o4 = a.loadObject(os.path.join(data_directory,
"roi/basal_ganglia.hie"))
test_assert(o4 is not None, 'object o4 is None')
print("o4 : nomenclature basal_ganglia.hie, internalRep = ", o4)
o3 = a.loadObject(os.path.join(data_directory,
"roi/basal_ganglia.arg"))
test_assert(o3 is not None, 'object o3 is None')
print("o3 : graph basal_ganglia.arg, internalRep = ", o3)
# this should fail
print(
'o5 : trying to load an object with wrong type (field restrict_object_type doesn\'t match the type of the object): it should fail.')
o5 = a.loadObject(filename=os.path.join(data_directory,
"subject01/subject01.nii"),
restrict_object_types={'Volume': ['FLOAT']})
# test_assert(o5 is None, 'object o5 is not None')
print('The error message is normal, o5 = ', o5)
o6 = a.loadObject(filename=os.path.join(data_directory,
"subject01/subject01.nii"),
restrict_object_types={'Volume': ['S16', 'FLOAT']})
test_assert(o6 is not None, 'object o6 is None')
print(
"o6 : volume subject01.ima, restrict_object_type match the object type. internalRep = ", o6)
o7 = a.loadObject(
filename=os.path.join(data_directory, "subject01/subject01.nii"))
test_assert(o7 is not None, 'object o7 is None')
print("o7 : same object")
cur = a.loadCursor(os.path.join(data_directory,
"subject01/subject01_Lhemi.mesh"))
test_assert(cur is not None, 'object cur is None')
print(
"cur : load Lhemi.mesh as a cursor. The object is not in the list of objects in Anatomist main window but can be selected as cursor in preferences. ")
test_assert(cur not in a.getObjects(),
'object cur is in Anatomist global list')
print("\n--- FusionObjects ---")
fus = a.fusionObjects([o, o2], "FusionTexSurfMethod", interactive)
test_assert(fus is not None, 'fusion fus is None')
print(
"fus : fusion of o and o2, method is FusionTexSurfMethod and askOrder is True, so a window opens to let the user choose the order of the objects in the fusion. internalRep = ", fus)
print("\n--- CreateReferential ---")
r = a.createReferential(os.path.join(
soma.config.BRAINVISA_SHARE,
"registration/Talairach-MNI_template-SPM.referential"))
test_assert(r is not None, 'referential r is None')
print(
"r : Referential loaded from the Talairach-MNI_template-SPM.referential. r.__class__ = ", r.__class__, ", internalRep = ", r, ", r.refUuid = ",
r.refUuid, ". This should not create a new referential because Talairach-MNI_template-SPM referential is already loaded in Anatomist. ")
test_assert(r == a.mniTemplateRef,
'referenrial r is not the MNI referential')
r2 = a.createReferential()
test_assert(r2 is not None, 'referential r2 is None')
print("r2 : new referential. internalRep = ", r2, ", refUuid = ",
r2.refUuid)
cr = a.centralRef
test_assert(cr is not None, 'central ref is None')
print("cr : central referential, ", cr, ", refUuid = ", cr.refUuid,
". This referential is already loaded in Anatomist.")
print("\n--- LoadTransformation ---")
t = a.loadTransformation(os.path.join(
data_directory,
"subject01/RawT1-subject01_default_acquisition_TO_Talairach-ACPC.trm"),
r2, cr)
test_assert(t is not None, 'transformation t is None')
print(
"t : loaded from the file chaos_TO_talairach.trm, as a transformation between r2 and cr. t.__class__ = ",
t.__class__, ", internalRep = ", t)
print("\n--- CreatePalette ---")
p = a.createPalette("maPalette")
test_assert(p is not None, 'palette p is None')
print(
"p : new palette named maPalette, added Anatomist list of palettes. p.__class__ = ",
p.__class__, ", internalRep = ", p)
print("\n--- GroupObjects ---")
g = a.groupObjects([o, o2])
test_assert(g is not None, 'group g is None')
print("g : new group of objects containing o and o2. g.__class__ = ",
g.__class__, ", internalRep = ", g)
test_assert(o in g.children and o2 in g.children,
'objects o or o2 are not children of group g')
print("\n--- linkWindows ---")
wg = a.linkWindows([w1, w2])
test_assert(wg is not None, 'window group wg is None')
print("wg : new group of windows containing w1 and w2. wg.__class__ = ",
wg.__class__, ", internalRep = ", wg)
print("\n--- GetInfos ---")
lo = a.getObjects()
test_assert(lo is not None, 'objects list lo is None')
print("\nObjects refererenced in current context : ", lo)
print("Total : ", len(lo))
nio = 79
if mode == "direct mode":
no = 79
nt = 6
nr = 3
else:
no = 9
nt = 1
# in socket mode r has a different ID as mniTemplateRef, but the same
# UUID, and it points to the same object in Anatomist. But we now
# see 4 refs from the client.
nr = 4
test_assert(len(lo) == no, 'wrong number of objects')
lio = a.importObjects(False) # top_level_only = False -> all objects
test_assert(lio is not None, 'objects list lio is None')
print(
"All objects (importing those that were not referenced in current context) : ", lio)
print("Total : ", len(lio))
print("-> Should be the same in direct implementation.")
test_assert(len(lio) == 79, 'wrong total number of objects')
lw = a.getWindows()
test_assert(lw is not None, 'windows list lw is None')
liw = a.importWindows()
test_assert(liw is not None, 'windows list liw is None')
print("\nGetWindows : ", len(lw), ", ImportWindows : ", len(liw))
print(liw)
test_assert(len(liw) == 5, 'wrong number of windows')
print("\nPalettes : ", a.getPalettes())
test_assert(len(a.getPalettes()) > 50, 'too few palettes')
lr = a.getReferentials()
test_assert(lr is not None, 'referentials list lr is None')
lir = a.importReferentials()
test_assert(lir is not None, 'referentials list lir is None')
print("\ngetReferentials : ", len(lr), ", importReferentials : ",
len(lir))
print(lir)
test_assert(len(lr) == nr, 'wrong referentials number: %d instead of %d'
% (len(lr), nr))
test_assert(len(lir) == nr,
'wrong imported referentials number: %d instead of %d'
% (len(lir), nr))
lt = a.getTransformations()
test_assert(lt is not None, 'transformations list lt is None')
lit = a.importTransformations()
test_assert(lit is not None, 'transformations list lit is None')
print("\ngetTransformations : ", len(lt), ", importTransformations : ",
len(lit))
test_assert(len(lt) == nt,
'wrong number of transformations: %d instead od %d'
% (len(lt), nt))
test_assert(len(lit) == 6,
'wrong number of imported transformations: %d instead of 6'
% len(lt))
sel = a.getSelection()
test_assert(sel is not None, 'selection sel is None')
print("\nSelections in default group", sel)
test_assert(len(sel) == 0, 'some objects are already selected')
print("\nCursor last pos", a.linkCursorLastClickedPosition())
print("Cursor last pos dans ref r2", a.linkCursorLastClickedPosition(r2))
print("\n--- AddObjects ---")
a.addObjects([fus], [w1, w2, w3])
print("Object fus added in windows w1, w2, and w3.")
test_assert(w1.objects == [fus], 'wrong objects in window w1')
print("\n--- RemoveObjects ---")
a.removeObjects([fus], [w2, w1])
print("Object fus removed from window w1 and w2.")
test_assert(len(w1.objects) == 0,
'objects still in window w1')
print("\n--- DeleteObjects ---")
# delete the list of objects to avoid keeping a reference on object that
# prevent from deleting it
del lo
del lio
a.deleteObjects([o7])
print("Delete object o7.")
test_assert(len(a.getObjects()) == nio - 1,
'wrong objects number after deletion of o7: %d instead of %d'
% (len(a.getObjects()), nio - 1))
print("\n--- AssignReferential ---")
a.assignReferential(r, [o2, w2])
print("Referential r assigned to object o2 and window w2.")
test_assert(w2.getReferential() == r, 'wrong referential in window w2')
test_assert(o2.referential is not None, 'No referential in object o6')
test_assert(o2.referential is not None and o2.referential == r,
'wrong referential in object o2: %s' % repr(o6.referential))
o6.assignReferential(r)
print(
"Referential r assigned to object o6. Should not have worked because o6 already has a transform to the MNI ref.")
test_assert(o6.referential is None or o6.referential != r,
'o6 referential has actually changed to r (wrong)')
print("\n--- camera ---")
a.camera([w3], zoom=1.5)
print("Set zoom to 1.5 in window w3.")
print("\n--- closeWindows ---")
# delete lists of windows to avoid keeping a reference that prevent from
# closing the window.
del lw
del liw
a.closeWindows([w4])
print("Close window w4.")
test_assert(len(a.getWindows()) == 4,
'wrong number of windows after deletion of w4: %d'
% len(a.getWindows()))
print("\n--- setMaterial ---")
o.addInWindows([w1])
mat = a.Material(diffuse=[0.5, 0.1, 0.1, 1], smooth_shading=1)
a.setMaterial([o], mat)
print("Add object o to the window w1 and change its material : ",
o.material)
test_assert(w1.objects == [o], 'wrong objects in w1')
test_assert(np.sum((np.array(o.getInfos()['material']['diffuse']) -
[0.5, 0.1, 0.1, 1]) ** 2) <= 1e-10,
'Material on o has not been updated correctly: %s'
% repr(o.getInfos()['material']['diffuse']))
print("\n--- setObjectPalette ---")
pal = a.getPalette("Blue-Red")
w6 = a.createWindow('Axial')
o6.addInWindows([w6])
a.setObjectPalette([o6], pal, minVal=0, maxVal=0.2)
print(
"Put object o6 in a new Axial window w6 and change its palette to Blue-Red with min and max values to 0 and 0.2")
test_assert(o6.getInfos()['palette']['palette'] == 'Blue-Red',
'Palette on o6 has not been set to "Blue-Red"')
print("\n--- setGraphParams ---")
a.setGraphParams(display_mode="mesh")
print("Set display mode (paint mode of objects in graph nodes) to mesh.")
print("\n--- AObject Methods---")
w7 = a.createWindow('3D')
o3.addInWindows([w7])
print("Put object o3 in new 3D window (w7). o3 attributes : filename : ",
o3.filename, ", material : ", o3.material, "objectType : ",
o3.objectType, ", children : ", o3.children)
o.addInWindows([w6])
o.removeFromWindows([w6])
print("\nAdd and remove object o from window w6.")
print(
"Try to delete o2. Should fail because the object is used in a fusion:")
o2.delete()
# Some methods are available in Anatomist class and Objects classes
# Anatomist.assignReferential -> AObject.assignReferential
# Anatomist.setMaterial -> AObject.setMaterial
# Anatomist.setObjectPalette -> AObject.setPalette
o4.assignReferential(r)
fus.setMaterial(mat)
o.setPalette(pal)
tex = fus.extractTexture()
test_assert(tex is not None, 'extracted texture is None')
print("\nExtract texture from object fus :", tex)
tex = fus.generateTexture()
test_assert(tex is not None, 'generated texture is None')
print("Generate a texture: tex =", tex)
fus2 = a.fusionObjects([o, tex], method="FusionTexSurfMethod")
fus2.addInWindows([w5])
print("Fusion the generated texture with object o : fus2 = ", fus2)
tmp_file = tempfile.mkstemp(suffix='.gii')
os.close(tmp_file[0])
fus.exportTexture(tmp_file[1])
a.sync() # wait for save operation to complete
test_assert(os.path.getsize(tmp_file[1]) != 0,
'saved texture file is empty')
print("fus texture is saved in file %s." % tmp_file[1])
tex = a.loadObject(tmp_file[1])
test_assert(tex is not None, 'could not re-read saved texture')
test_assert(tex.objectType == 'TEXTURE',
'loaded texture is not a texture...')
os.unlink(tmp_file[1])
os.unlink(tmp_file[1] + '.minf')
tmp_file = tempfile.mkstemp(suffix='.gii')
os.close(tmp_file[0])
o.save(tmp_file[1])
print("The object o is saved in the file %s." % tmp_file[1])
a.sync() # wait for save operation to complete
test_assert(os.path.getsize(tmp_file[1]) != 0, 'saved mesh file is empty')
mesh = a.loadObject(tmp_file[1])
test_assert(mesh is not None, 'could not re-read saved mesh')
test_assert(mesh.objectType == 'SURFACE', 'loaded mesh is not a mesh...')
os.unlink(tmp_file[1])
os.unlink(tmp_file[1] + '.minf')
print("\n--- AWindow Methods---")
print(
"Window attributes: w2.windowType = ", w2.windowType, ", w2.group = ",
w2.group)
test_assert(w2.windowType == 'Sagittal', 'w2 type is not Sagittal')
test_assert(w2.group == wg, 'w2 group is not wg')
# Some methods available in Anatomist class are also available directly in
# AWindows class.
w2.addObjects([o])
w2.removeObjects([o])
w2.camera(2)
w2.assignReferential(cr)
w2.addObjects([fus])
w2.moveLinkedCursor([150, 100, 60])
w6.showToolbox()
# opens the toolbox window. This toolbox will be empty if
# the window is empty. If there is an object on which it is
# possible to draw a roi, the roi toolbox will be shown.
print("\n--- AWindowsGroup Methods---")
wg.setSelection([fus])
print("Set fus object as selected in the group of windows wg : ")
print("-> selection in default group has not changed :", a.getSelection())
print("-> selection in the group of window wg :", a.getSelection(wg))
wg.unSelect([fus])
print("After unselect, selection in wg :", a.getSelection(wg))
g0 = a.getDefaultWindowsGroup() # the default group has the id 0
g0.setSelectionByNomenclature(o4, ['Caude_droit'])
print("Selection by nomenclature in default group - add 'Caude_droit':",
g0.getSelection())
g0.addToSelectionByNomenclature(o4, ['Caude_gauche'])
print("Selection by nomenclature default group - add 'Caude_gauche' :",
g0.getSelection())
g0.toggleSelectionByNomenclature(o4, ['Caude_gauche'])
print(
"Toggle selection by nomenclature in default group - toggle 'Caude_gauche' : ",
g0.getSelection())
print("\n--- APalette Methods---")
# set colors take as parameter a list of RGB components for colors :
# [r1,g1,b1,r2,g2,b2...]
p.setColors(colors=[100, 0, 0] * 20 + [0, 100, 0] * 20 + [0, 0, 100] * 20)
print("The colors of palette 'maPalette' has changed.")
print("\n--- ATransformation Methods---")
tmp_file = tempfile.mkstemp(suffix='.trm')
os.close(tmp_file[0])
t.save(tmp_file[1])
a.sync() # wait for save operation to complete
print("Save the transformation t in file %s." % tmp_file[1])
test_assert(os.path.getsize(tmp_file[1]) != 0,
'saved transformation file is empty')
os.unlink(tmp_file[1])
os.unlink(tmp_file[1] + '.minf')
# return objects and windows to keep a reference on them and avoid their
# destroying.
objects = a.getObjects()
windows = a.getWindows()
return (objects, windows)
def testBase():
# base module : simple interface, methods are not implemented
import anatomist.base as pyanatomist
a = pyanatomist.Anatomist()
w = a.createWindow('Axial')
print(w)
interactive = True
if len(sys.argv) >= 2 and "-b" in sys.argv[1:] or "--batch" in sys.argv[1:] \
or 'sphinx_gallery' in sys.modules:
interactive = False
print("\n**** TEST ANATOMIST API DIRECT IMPLEMENTATION ****\n")
res1 = testDirectImpl(interactive)
if not interactive:
del res1
print("\n**** TEST ANATOMIST API SOCKET IMPLEMENTATION ****\n")
res2 = testSocketImpl(interactive)
if not interactive:
del res2
if len(failures) != 0:
print('\n\n** tests have failed: **')
print('\n'.join(failures))
print()
raise RuntimeError('tests have failed:')
else:
print('\nTests OK.')
Using OpenGL in PyAnatomist¶
Customizing OpenGl parameters for objects
Download source: customopenglobject.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Using OpenGL in PyAnatomist
---------------------------
Customizing OpenGL parameters for objects
'''
from soma import aims
import anatomist.direct.api as anatomist
from soma.qt_gui.qt_backend import Qt
from OpenGL import GL
import six
import sys
class VolRender(anatomist.cpp.ObjectVector, anatomist.cpp.GLComponent):
def __init__(self, objects):
self._objects = objects
for o in objects:
self.insert(o)
def renderingIsObserverDependent(self):
return True
class WinViewMesh (anatomist.cpp.ASurface_3):
def __init__(self, mesh, followorientation=True, followposition=False):
if mesh is not None:
if type(mesh) is six.string_types:
# mesh is a filename: read it
anatomist.cpp.ASurface_3.__init__(self, mesh)
r = aims.Reader()
m = aims.rc_ptr_AimsTimeSurface_3(r.read(mesh))
self.setSurface(m)
else:
# mesh should be an Aims mesh: assign it
anatomist.cpp.ASurface_3.__init__(self)
self.setSurface(mesh)
else:
# generate a sphere mesh
anatomist.cpp.ASurface_3.__init__(self)
m = aims.rc_ptr_AimsTimeSurface_3(aims.SurfaceGenerator.sphere(
aims.Point3df(0, 0, 0), 10, 100))
self.setSurface(m)
self._followorientation = followorientation
self._followposition = followposition
def renderingIsObserverDependent(self):
return True
def glMainGLL(self, state):
self.glSetChanged(anatomist.cpp.GLComponent.glGENERAL)
return anatomist.cpp.GLComponent.glMainGLL(self, state)
def glBeforeBodyGLL(self, state, prim):
if self._followorientation and self._followposition:
return
gll = anatomist.cpp.GLList()
gll.generate()
prim.append(gll)
GL.glNewList(gll.item(), GL.GL_COMPILE)
GL.glMatrixMode(GL.GL_MODELVIEW)
GL.glPushMatrix()
GL.glMatrixMode(GL.GL_PROJECTION)
GL.glPushMatrix()
if not self._followposition:
winDim = 70
GL.glPushAttrib(GL.GL_VIEWPORT_BIT)
GL.glViewport(0, 0, winDim, winDim)
GL.glLoadIdentity()
orthoMinX = - 1.5
orthoMinY = - 1.5
orthoMinZ = - 1.5
orthoMaxX = 1.5
orthoMaxY = 1.5
orthoMaxZ = 1.5
GL.glOrtho(orthoMinX, orthoMaxX, orthoMinY, orthoMaxY,
orthoMinZ, orthoMaxZ)
win = state.window
if win \
and isinstance(win, anatomist.cpp.ControlledWindow):
view = win.view()
if not self._followorientation:
GL.glTranslate(1, 1, 0)
# mat = GL.glGetFloatv( GL.GL_MODELVIEW_MATRIX )
# scale = aims.Point3df( mat[0][0], mat[1][0], mat[2][0] ).norm()
GL.glMatrixMode(GL.GL_MODELVIEW)
GL.glLoadIdentity()
# keep the translation part of the view orientation
# (mut apply the inverse rotation to it)
trans = view.rotationCenter()
r = aims.AffineTransformation3d(view.quaternion()).inverse()
r = r * aims.AffineTransformation3d([1, 0, 0, trans[0],
0, 1, 0, trans[1], 0, 0, 1, -trans[2], 0, 0, 0, 1])
GL.glTranslate(-r.translation()[0], -r.translation()[1],
-r.translation()[2])
GL.glScalef(1., 1., -1.)
GL.glMatrixMode(GL.GL_PROJECTION)
else:
GL.glMatrixMode(GL.GL_MODELVIEW)
# keep the rotation part of the view orientation, removing the
# translation part
trans = view.rotationCenter()
r = aims.AffineTransformation3d(view.quaternion()).inverse()
r = aims.AffineTransformation3d([1, 0, 0, trans[0],
0, 1, 0, trans[1], 0, 0, 1, trans[2], 0, 0, 0, 1])
GL.glTranslate(r.translation()[0], r.translation()[1],
r.translation()[2])
GL.glMatrixMode(GL.GL_PROJECTION)
GL.glEndList()
def glAfterBodyGLL(self, state, prim):
if self._followorientation and self._followposition:
return
gll = anatomist.cpp.GLList()
gll.generate()
prim.append(gll)
GL.glNewList(gll.item(), GL.GL_COMPILE)
GL.glMatrixMode(GL.GL_PROJECTION)
if not self._followposition:
GL.glPopAttrib(GL.GL_VIEWPORT_BIT)
GL.glPopMatrix()
GL.glMatrixMode(GL.GL_MODELVIEW)
GL.glPopMatrix()
GL.glEndList()
# ---
if __name__ == '__main__':
runloop = False
if Qt.QApplication.instance() is None and 'IPython' not in sys.modules:
runloop = True
a = anatomist.Anatomist()
mesh = aims.SurfaceGenerator.sphere((0, 0, 0), 1., 200)
amesh = anatomist.cpp.AObjectConverter.anatomist(mesh)
cube1 = aims.SurfaceGenerator.cube((0, 0, 0), 0.5, False)
vcube1 = WinViewMesh(cube1)
a.registerObject(vcube1)
cube2 = aims.SurfaceGenerator.cone((0, -0.5, 0), (0, 0.5, 0), 0.5, 50,
True, False)
vcube2 = WinViewMesh(cube2, followorientation=False,
followposition=True)
a.registerObject(vcube2)
w = a.createWindow('3D')
a.addObjects([amesh, vcube1, vcube2], [w])
import sys
if 'sphinx_gallery' in sys.modules:
# display in matplotlib for sphinx_gallery
import matplotlib
w.camera(view_quaternion=[0.535079479217529,
0.797160744667053,
0.268512755632401,
0.0782672688364983],
zoom=0.45)
matplotlib.use('agg', force=True) # force agg
w.imshow(show=True)
runloop = False
if runloop:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del w, amesh, vcube1, vcube2, cube1, cube2, mesh
Texture drawing¶
Drawing ROI on a mesh in a texture
Download source: texturedrawing.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Texture drawing
---------------
Drawing ROI on a mesh in a texture. Of course the SurfPaint module allows to do it with much richer features, but this example demonstrates how to code the basics of it in a relatively small program.
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
from soma.aims import colormaphints
import sys
import os
import math
import six
sys.path.insert(0, '.')
userLevel = 4
# determine wheter we are using Qt4 or Qt5, and hack a little bit accordingly
# the boolean qt4 gloabl variable will tell it for later usage
from soma.qt_gui.qt_backend import QtCore, QtGui
from soma.qt_gui.qt_backend import loadUi
# do we have to run QApplication ?
if QtGui.QApplication.instance() is None:
runqt = True
else:
runqt = False
import sphere
import numpy
selmesh = None
selanamesh = None
class TexDrawAction(anatomist.cpp.Action):
def name(self):
return 'TexDrawAction'
def takePolygon(self, x, y, globx, globy):
# print('takePolygon', x, y)
w = self.view().aWindow()
obj = w.objectAtCursorPosition(x, y)
if obj is not None:
print('object:', obj, obj.name())
surf = None
if obj.objectTypeName(obj.type()) == 'SURFACE':
surf = obj
elif isinstance(obj, anatomist.cpp.MObject):
surf = [o for o in obj if o.type() == 3]
if len(surf) != 1:
return
surf = surf[0]
if surf is not None:
poly = w.polygonAtCursorPosition(x, y, obj)
if poly == 0xffffff: # white
return # background
print('polygon:', poly)
mesh = anatomist.cpp.AObjectConverter.aims(surf)
# print('mesh:', mesh)
ppoly = mesh.polygon()[poly]
vert = mesh.vertex()
global selmesh, selanamesh
if selmesh is None:
selmesh = aims.AimsSurfaceTriangle()
selmesh.vertex().assign([vert[ppoly[0]], vert[ppoly[1]],
vert[ppoly[2]]])
selmesh.polygon().assign([aims.AimsVector_U32_3(0, 1, 2)])
if selanamesh is None:
selanamesh = anatomist.cpp.AObjectConverter.anatomist(
selmesh)
a = anatomist.Anatomist()
a.execute('SetMaterial', objects=[
selanamesh], diffuse=[0, 0, 1., 1.])
a.execute('AddObject', objects=[selanamesh], windows=[w])
selanamesh.setChanged()
selanamesh.notifyObservers()
def delPolygon(self):
global selmesh, selanamesh
selmesh = None
# keep object ID and release python reference to it
id = a.convertSingleObjectParamsToIDs(selanamesh)
selanamesh = None
a.execute('DeleteElement', elements=[id])
def newtexture(self, x, y, globx, globy):
print('new texture')
w = self.view().aWindow()
aw = a.AWindow(a, w)
obj = w.objectAtCursorPosition(x, y)
# print('object:', obj)
if obj is not None:
if obj.objectTypeName(obj.type()) == 'SURFACE':
surf = obj
texs = []
# elif obj.objectTypeName( obj.type() ) == 'TEXTURED SURF.':
# print('TEXTURED SURF.')
# surf = [ o for o in obj if o.type() == 3 ]
# if len( surf ) != 1:
# print('not one mesh, but', len( surf ))
# return
# surf = surf[0]
# texs = [ o for o in obj if o.type() == 18 ]
# self._texsurf = obj
# print('draw initiated')
# return
else:
return
gl = surf.glAPI()
if gl:
vs = anatomist.cpp.ViewState()
nv = gl.glNumVertex(vs)
if nv > 0:
tex = aims.TimeTexture_FLOAT()
t = tex[0]
t.reserve(nv)
for i in six.moves.xrange(nv):
t.push_back(0.)
atex = a.toAObject(tex)
atex.releaseAppRef()
# atex = a.AObject( a, surf ).generateTexture()
texs.append(atex)
# ...
tsurf = a.fusionObjects(
[atex, obj], method='FusionTexSurfMethod')
tsurf.setPalette(palette='Blue-Red-fusion')
self._texsurf = tsurf
# tsurf.takeRef()
aw.removeObjects(obj)
aw.addObjects(tsurf)
def startDraw(self, x, y, globx, globy):
self._startDraw(x, y, 1.)
def startErase(self, x, y, globx, globy):
self._startDraw(x, y, 0.)
def _startDraw(self, x, y, value):
w = self.view().aWindow()
obj = w.objectAtCursorPosition(x, y)
# print('object:', obj)
if obj is not None:
texs = []
if obj.objectTypeName(obj.type()) == 'SURFACE':
surf = obj
p = obj.parents()
found = False
for o in p:
if o.objectTypeName(o.type()) == 'TEXTURED SURF.' \
and w.hasObject(o):
texs = [ob for ob in o if ob.type() == 18]
self._texsurf = o
found = True
break
if not found:
# create a new texture
self.newtexture(x, y, 0, 0)
self._startDraw(x, y, value)
return
elif obj.objectTypeName(obj.type()) == 'TEXTURED SURF.':
surf = [o for o in obj if o.type() == 3]
if len(surf) != 1:
return
surf = surf[0]
texs = [o for o in obj if o.type() == 18]
self._texsurf = obj
else:
return
if len(texs) == 0:
return
self._surf = surf
self._tex = texs[-1]
self._mesh = anatomist.cpp.AObjectConverter.aims(surf)
self._aimstex = anatomist.cpp.AObjectConverter.aims(
self._tex, {'scale': 0})
self.draw(x, y, value)
def endDraw(self, x, y, globx, globy):
if hasattr(self, '_aimstex'):
del self._aimstex
del self._mesh
del self._tex
del self._surf
def moveDraw(self, x, y, globx, globy):
self.draw(x, y, 1.)
def erase(self, x, y, globx, globy):
self.draw(x, y, 0.)
def draw(self, x, y, value):
if not hasattr(self, '_aimstex'):
return
w = self.view().aWindow()
obj = self._texsurf
poly = w.polygonAtCursorPosition(x, y, obj)
if poly == 0xffffff or poly < 0 or poly >= len(self._mesh.polygon()):
return
ppoly = self._mesh.polygon()[poly]
print('poly:', poly, ppoly)
vert = self._mesh.vertex()
pos = aims.Point3df()
pos = w.positionFromCursor(x, y)
print('pos:', pos)
v = ppoly[numpy.argmin([(vert[p] - pos).norm() for p in ppoly])]
print('vertex:', v, vert[v])
self._aimstex[0][v] = value
self._tex.setChanged()
self._tex.notifyObservers()
class TexDrawControl(anatomist.cpp.Control):
def __init__(self, prio=25):
anatomist.cpp.Control.__init__(self, prio, 'TexDrawControl')
def eventAutoSubscription(self, pool):
key = QtCore.Qt
NoModifier = key.NoModifier
ShiftModifier = key.ShiftModifier
ControlModifier = key.ControlModifier
AltModifier = key.AltModifier
self.mouseLongEventSubscribe(
key.LeftButton, NoModifier,
pool.action('TexDrawAction').startDraw,
pool.action('TexDrawAction').moveDraw,
pool.action('TexDrawAction').endDraw,
False)
self.mouseLongEventSubscribe(
key.LeftButton, NoModifier,
pool.action('TexDrawAction').startDraw,
pool.action('TexDrawAction').moveDraw,
pool.action('TexDrawAction').endDraw,
False)
self.mouseLongEventSubscribe(
key.LeftButton, ControlModifier,
pool.action('TexDrawAction').startErase,
pool.action('TexDrawAction').erase,
pool.action('TexDrawAction').endDraw,
False)
self.mousePressButtonEventSubscribe(
key.RightButton, ControlModifier,
pool.action('TexDrawAction').newtexture)
# polygon picking
self.mousePressButtonEventSubscribe(key.RightButton, NoModifier,
pool.action('TexDrawAction').takePolygon)
self.keyPressEventSubscribe(key.Key_Escape, NoModifier,
pool.action("TexDrawAction").delPolygon)
# now plug the standard actions
self.mouseLongEventSubscribe(key.MidButton, ShiftModifier,
pool.action("Zoom3DAction").beginZoom,
pool.action("Zoom3DAction").moveZoom,
pool.action("Zoom3DAction").endZoom, True)
self.wheelEventSubscribe(pool.action("Zoom3DAction").zoomWheel)
self.keyPressEventSubscribe(key.Key_C, ControlModifier,
pool.action("Trackball").setCenter)
self.keyPressEventSubscribe(key.Key_C, AltModifier,
pool.action("Trackball").showRotationCenter)
self.mouseLongEventSubscribe(key.MidButton, ControlModifier,
pool.action(
"Translate3DAction").beginTranslate,
pool.action(
"Translate3DAction").moveTranslate,
pool.action("Translate3DAction").endTranslate, True)
self.mouseLongEventSubscribe(
key.MidButton, NoModifier,
pool.action('ContinuousTrackball').beginTrackball,
pool.action('ContinuousTrackball').moveTrackball,
pool.action('ContinuousTrackball').endTrackball, True)
self.keyPressEventSubscribe(key.Key_Space, ControlModifier,
pool.action("ContinuousTrackball").startOrStop)
# a = anatomist.Anatomist()
# pix = QtGui.QPixmap( 'control.xpm' )
# anatomist.cpp.IconDictionary.instance().addIcon( 'MyControl',
# pix )
# ad = anatomist.cpp.ActionDictionary.instance()
# ad.addAction( 'MyAction', lambda: MyAction() )
# cd = anatomist.cpp.ControlDictionary.instance()
# cd.addControl( 'MyControl', lambda: MyControl(), 25 )
# cm = anatomist.cpp.ControlManager.instance()
# cm.addControl( 'QAGLWidget3D', '', 'MyControl' )
# s = sphere.ASphere()
# a.registerObject( s )
# aw = a.createWindow( '3D' )
# a.addObjects( [ s ], [ aw ] )
a = anatomist.Anatomist()
qapp = QtGui.QApplication.instance()
pix = QtGui.QPixmap('control.xpm')
anatomist.cpp.IconDictionary.instance().addIcon('TexDrawControl',
pix)
ad = anatomist.cpp.ActionDictionary.instance()
ad.addAction('TexDrawAction', TexDrawAction)
cd = anatomist.cpp.ControlDictionary.instance()
cd.addControl('TexDrawControl', TexDrawControl, 26)
cm = anatomist.cpp.ControlManager.instance()
cm.addControl('QAGLWidget3D', '', 'TexDrawControl')
s = a.loadObject('test.mesh')
aw = a.createWindow('3D')
a.addObjects([s], [aw])
a.execute('SetControl', windows=[aw], control='TexDrawControl')
QtGui.QMessageBox.information(
None, 'texture drawing', '1. put a mesh in a 3D view\n2.select the "Mickey" control\n3. ctrl+right click on the mesh to create an empty texture or initiate the drawing session\n4. draw on the mesh using the mouse left button\n ctrl+left button erases', QtGui.QMessageBox.Ok)
# run Qt
if runqt:
qapp.exec_()
del ad, cm, aw, s, pix
Measurements on a volume¶
Complete example with a specific graphical interface that embeds an Anatomist window and a matplotlib widget. Shows a 4D volume and a curve of the values in a selected voxel or the mean of values in a selected region.
Download source: volumeMeasures.py
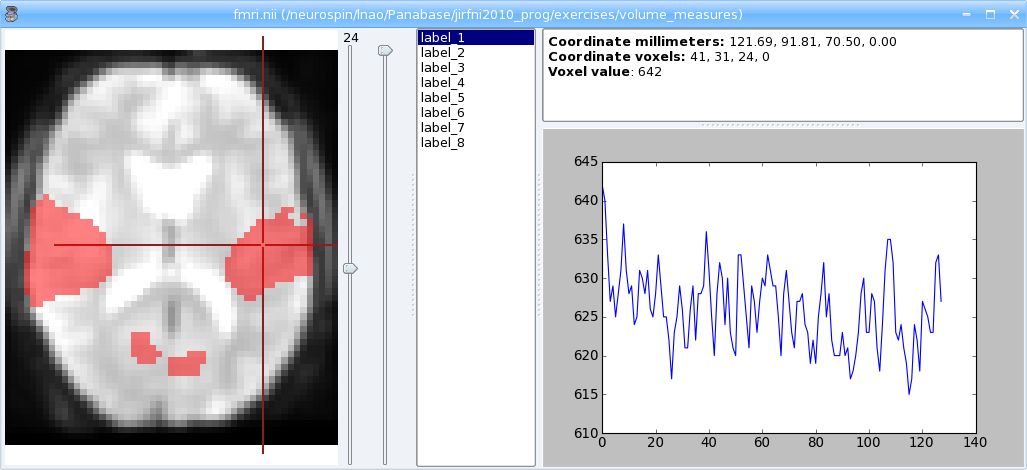
Volume Measures Example¶
#! /bin/env python
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Measurements on a volume
------------------------
Complete example with a specific graphical interface that embeds an Anatomist window and a matplotlib widget. Shows a 4D volume and a curve of the values in a selected voxel or the mean of values in a selected region.
'''
from __future__ import absolute_import
import sys
import os
import weakref
import gc
import operator
import anatomist.direct.api as anatomist
from soma.qt_gui.qt_backend.QtCore import Qt, QPoint
from soma.qt_gui.qt_backend.QtGui import QSplitter, QListWidget, QTextEdit, \
QApplication
from soma import aims
from soma.qt_gui import qt_backend
import six
from six.moves import zip
from functools import reduce
qt_backend.init_matplotlib_backend()
import matplotlib
import numpy
import pylab
FigureCanvas = qt_backend.FigureCanvas
from matplotlib.figure import Figure
class MeasuresWindow(QSplitter):
def __init__(self, fileName, roiIterator=None, parent=None,
anatomistInstance=None):
QSplitter.__init__(self, Qt.Horizontal, parent)
if anatomistInstance is None:
# initialize Anatomist
self.anatomist = anatomist.Anatomist()
else:
self.anatomist = anatomistInstance
# open an axial window
self.aWindow = self.anatomist.createWindow(
'Axial', no_decoration=True)
self.aWindow.setParent(self)
if roiIterator is not None:
self.roiList = QListWidget(self)
self.maskIterators = []
# Iterate on each region
while roiIterator.isValid():
self.roiList.addItem(roiIterator.regionName())
maskIterator = roiIterator.maskIterator().get()
maskIterator.bucket = None
self.maskIterators.append(maskIterator)
next(roiIterator)
self.selectedBucket = None
self.roiList.currentRowChanged.connect(self.regionSelected)
else:
self.roiList = None
self.infoSplitter = QSplitter(Qt.Vertical, self)
self.info = QTextEdit(self.infoSplitter)
self.info.setReadOnly(True)
self.matplotFigure = Figure()
self.matplotAxes = self.matplotFigure.add_subplot(111)
# We want the axes cleared every time plot() is called
self.matplotAxes.hold(False)
self.matplotCanvas = FigureCanvas(self.matplotFigure)
self.matplotCanvas.setParent(self.infoSplitter)
self.matplotCanvas.updateGeometry()
self.anatomist.onCursorNotifier.add(self.clicked2)
self.resize(800, 600)
# Read the image
dir, base = os.path.split(fileName)
if dir:
self.setWindowTitle(base + ' (' + dir + ')')
else:
self.setWindowTitle(base)
# load any volume as a aims.Volume_* object
self.volume = aims.read(fileName)
self.interpolator = aims.aims.getLinearInterpolator(self.volume).get()
# convert the carto volume to Anatomist API
avol = self.anatomist.toAObject(self.volume)
avol.releaseAppRef()
# put volume in window
self.aWindow.addObjects(avol)
self._ignoreClicked = False
voxelSize = self.volume.header()['voxel_size']
tmp = self.volume.header()['volume_dimension']
volumeSize = [int(i) for i in tmp]
volumeCenter = [v * s / 2 for v, s in zip(volumeSize, voxelSize)]
self.clicked(volumeCenter)
infoHeight = self.info.sizeHint().height()
self.infoSplitter.setSizes([infoHeight, self.height() - infoHeight])
def clicked2(self, eventName, eventParameters):
self.clicked(eventParameters['position'], eventParameters['window'])
def clicked(self, posMM, aWindow=None):
posMM = [float(i) for i in posMM]
if self._ignoreClicked:
return
text = '<html><body>\n'
text += '<b>Coordinate millimeters:</b> %.2f, %.2f, %.2f, %.2f' % tuple(
posMM) + '<br/>\n'
voxelSize = self.volume.header()['voxel_size']
posVoxel = [int(round(i / j)) for i, j in zip(posMM, voxelSize)]
text += '<b>Coordinate voxels:</b> %d, %d, %d, %d' % tuple(
posVoxel) + '<br/>\n'
tmp = self.volume.header()['volume_dimension']
volumeSize = [int(i) for i in tmp]
if not [None for i in posVoxel if i < 0] and \
not [None for i, j in zip(posVoxel, volumeSize) if i >= j]:
text += '<b>Voxel value</b>: ' + \
str(self.volume.value(*posVoxel)) + '<br/>\n'
if volumeSize[3] > 1:
indices = numpy.arange(volumeSize[3])
# Extract values as numarray structure
values = self.interpolator.values(posVoxel[0] * voxelSize[0],
posVoxel[1] * voxelSize[1],
posVoxel[2] * voxelSize[2])
self.matplotAxes.plot(indices, numpy.array(values))
self.matplotCanvas.draw()
text += '</body></html>'
self.info.setText(text)
def regionSelected(self):
index = self.roiList.currentRow()
if index >= 0:
text = '<html><body>\n'
text += '<h2>' + \
six.text_type(self.roiList.item(index).text()) + '</h2>\n'
maskIterator = self.maskIterators[index]
if maskIterator.bucket is None:
roiCenter = aims.Point3df(0, 0, 0)
bucket = aims.BucketMap_VOID()
bucket.setSizeXYZT(
*list(maskIterator.voxelSize().items()) + (1,))
maskIterator.restart()
valid = 0
invalid = 0
sum = None
# Iterate on each point of a region
while maskIterator.isValid():
bucket[0][maskIterator.value()] = 1
p = maskIterator.valueMillimeters()
roiCenter += p
# Check if the point is in the image limit
if self.interpolator.isValid(p):
values = self.interpolator.values(p)
if sum is None:
sum = values
else:
sum = [s + v for s, v in zip(sum, values)]
valid += 1
else:
invalid += 1
next(maskIterator)
text += '<b>valid points:</b> ' + str(valid) + '<br/>\n'
text += '<b>invalid points:</b> ' + str(invalid) + '<br/>\n'
if valid:
means = [s / float(valid) for s in sum]
mean = reduce(operator.add, means) / len(means)
else:
means = []
mean = 'N/A'
text += '<b>mean:</b> ' + str(mean) + '<br/>\n'
text += '</body></html>'
maskIterator.text = text
# convert the BucketMap to Anatomist API
maskIterator.bucket = self.anatomist.toAObject(bucket)
maskIterator.bucket.releaseAppRef()
maskIterator.bucket.setName(
str(self.roiList.item(index).text()))
maskIterator.bucket.setChanged()
count = valid + invalid
if count:
maskIterator.roiCenter = [
c / count for c in roiCenter.items()]
else:
maskIterator.roiCenter = None
maskIterator.means = means
# put bucket in window
self.info.setText(maskIterator.text)
self.aWindow.addObjects([maskIterator.bucket])
# Set selected color to bucket
maskIterator.bucket.setMaterial(self.anatomist.Material(
diffuse=[1, 0, 0, 0.5],
lighting=0,
face_culling=1,
))
# Set unselected color to previously selected bucket
if self.selectedBucket is not None:
self.selectedBucket.setMaterial(
self.anatomist.Material(diffuse=[0, 0.8, 0.8, 0.8]))
self.selectedBucket = maskIterator.bucket
if maskIterator.roiCenter is not None:
self._ignoreClicked = True
self.aWindow.moveLinkedCursor(maskIterator.roiCenter)
self._ignoreClicked = False
if len(maskIterator.means) > 1:
indices = numpy.arange(len(maskIterator.means))
self.matplotAxes.plot(
indices, numpy.array(maskIterator.means))
self.matplotCanvas.draw()
if __name__ == '__main__':
qApp = QApplication(sys.argv)
if len(sys.argv) == 3:
roiIterator = aims.aims.getRoiIterator(sys.argv[2]).get()
w = MeasuresWindow(sys.argv[1], roiIterator=roiIterator)
else:
w = MeasuresWindow(sys.argv[1])
w.show()
anatomist.Anatomist().getControlWindow().hide()
qApp.exec_()
del w
3D Text annotations on a graph¶
Display graph labels as 3D text annotations.
Download source: anagraphannotate.py
#!/usr/bin/env python
# -*- coding: utf-8 -*-
'''
3D Text annotations on a graph
==============================
Display graph labels as 3D text annotations. This code is somewhat equivalent to what happens using the 'A' key in selection control mode (actually this is an ancestor of the module which has been incorporated in Anatomist).
'''
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as ana
from soma import aims
import os
import numpy
import sys
from soma.qt_gui.qt_backend import QtGui
import six
byvertex = False
class Props(object):
def __init__(self):
self.lvert = []
self.lpoly = []
self.usespheres = True
self.colorlabels = True
self.center = aims.Point3df()
def makelabel(label, gc, pos, color, props):
objects = []
to = ana.cpp.TextObject(label)
to.setScale(0.1)
to.setName('label: ' + label)
a.registerObject(to, False)
a.releaseObject(to)
if label in colors:
color = colors[label]
if props.usespheres:
sph = aims.SurfaceGenerator.icosphere(gc, 2, 50)
asph = a.toAObject(sph)
asph.setMaterial(diffuse=color)
asph.setName('gc: ' + label)
a.unregisterObject(asph)
a.registerObject(asph, False)
a.releaseObject(asph)
objects.append(asph)
if props.colorlabels:
to.GetMaterial().set({'diffuse': color})
texto = ana.cpp.TransformedObject([to], False, True, pos)
texto.setDynamicOffsetFromPoint(props.center)
texto.setName('annot: ' + label)
objects.append(texto)
props.lpoly.append(aims.AimsVector_U32_2((len(props.lvert),
len(props.lvert) + 1)))
props.lvert += [gc, pos]
a.registerObject(texto, False)
a.releaseObject(texto)
return objects
runloop = False
if QtGui.QApplication.startingUp() and 'IPython' not in sys.modules:
runloop = True
a = ana.Anatomist()
qapp = QtGui.QApplication.instance()
share = aims.carto.Paths.globalShared()
nomenclname = os.path.join(aims.carto.Paths.shfjShared(), 'nomenclature',
'hierarchy', 'sulcal_root_colors.hie')
graphname = os.path.join(share, 'doc', 'pyanatomist-' + '.'.join(
[str(x) for x in aims.version()]), 'examples', 'Rbase.arg')
labelatt = 'name'
nomenclature = a.loadObject(nomenclname)
graph = aims.read(graphname)
if 'label_property' in graph:
labelatt = graph['label_property']
agraph = a.toAObject(graph)
agraph.releaseAppRef()
w = a.createWindow('3D')
w.addObjects(agraph, add_graph_nodes=True)
# lgraphaims = aims.Graph( 'labelsGraph' )
# lgraph = a.toAObject( lgraphaims )
bbox = agraph.boundingbox()
bbox = (aims.Point3df(bbox[0][:3]), aims.Point3df(bbox[1][:3]))
props = Props()
props.center = (bbox[0] + bbox[1]) / 2
size = (bbox[1] - bbox[0]).norm() * 0.2
vs = graph['voxel_size'][:3]
objects = []
lines = aims.TimeSurface(2)
elements = {}
colors = {}
for v in graph.vertices():
if 'gravity_center' in v and labelatt in v:
gc = aims.Point3df(numpy.array(v['gravity_center']) * vs)
label = v[labelatt]
if label != 'unknown':
if label not in elements:
elem = [aims.Point3df(0, 0, 0), 0.]
elements[label] = elem
else:
elem = elements[label]
sz = v['size']
elem[0] += gc * sz
elem[1] += sz
color = [0, 0, 0, 1]
if 'ana_object' in v:
av = v['ana_object']
color = av.GetMaterial().genericDescription()['diffuse']
colors[label] = color
if byvertex:
pos = gc + (gc - props.center).normalize() * size
objects += makelabel(label, gc, pos, color, props)
if not byvertex:
for label, elem in six.iteritems(elements):
gc = elem[0] / elem[1]
pos = gc + (gc - props.center).normalize() * size
if label in colors:
color = colors[label]
else:
color = [0, 0, 0, 1]
objects += makelabel(label, gc, pos, color, props)
lines.vertex().assign(props.lvert)
lines.polygon().assign(props.lpoly)
alines = a.toAObject(lines)
a.unregisterObject(alines)
a.registerObject(alines, False)
a.releaseObject(alines)
del lines
alines.setMaterial(diffuse=[0, 0, 0, 1])
objects.append(alines)
del alines
labels = a.groupObjects(objects)
labels.releaseAppRef()
w.addObjects(labels, add_children=True)
if 'sphinx_gallery' in sys.modules:
w.windowConfig(view_size=[907, 568])
w.camera(view_quaternion=[0.520213842391968,
-0.42516353726387,
-0.470633894205093,
0.571941733360291],
zoom=0.8187)
# display in matplotlib for sphinx_gallery
w.sphinx_gallery_snapshot()
runloop = False
if runloop:
qapp.exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del w, nomenclature, agraph, graph, objects, labels, elements, colors
Activating actions in the current control¶
Activate actions in the current contol of a window
Download source: activateaction.py
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Action activation triggering API demo
=====================================
Activate actions in the current control of a window.
Action activation works both in direct and socket APIs
but querying the lists of available action methods is only available in
direct mode.
'''
import anatomist.direct.api as anatomist
from soma.qt_gui.qt_backend import Qt
import sys
runloop = False
if Qt.QApplication.instance() is None:
runloop = True
a = anatomist.Anatomist()
vol = a.loadObject('irm.ima')
w = a.createWindow('Axial')
w.addObjects(vol)
w.activateAction(action_type='key_press', method='movie_start_stop')
# set backward movie mode
w.activateAction(action_type='key_press', method='movie_next_mode')
# loop forward
w.activateAction(action_type='key_press', method='movie_next_mode')
# loop backward
w.activateAction(action_type='key_press', method='movie_next_mode')
# loop both ways
w.activateAction(action_type='key_press', method='movie_next_mode')
# query available actions methods
c = w.view().controlSwitch().activeControlInstance()
print('in control:', c.name())
kpmethods = c.keyPressActionLinkNames()
print('* keyPress methods:', kpmethods)
mpmethods = c.mousePressActionLinkNames()
print('* mousePress methods:', mpmethods)
w.setControl('SelectionControl')
c = w.view().controlSwitch().activeControlInstance()
print('in control:', c.name())
kpmethods = c.keyPressActionLinkNames()
print('* keyPress methods:', kpmethods)
mpmethods = c.mousePressActionLinkNames()
print('* mousePress methods:', mpmethods)
if runloop: # and 'sphinx_gallery' not in sys.modules:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del c, w, vol, kpmethods, mpmethods
Simple viewer¶
An even simplified version of the anasimpleviewer application, which may also be used as a programming example. Its code is in the “bin/”” directory of the binary packages.
Download source: anaevensimplerviewer.py
Ana Even Simpler Viewer Example¶
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Build a custom simplified viewer based on Anatomist
===================================================
An even simplified version of the anasimpleviewer application, which may also
be used as a programming example. Its code is in the “bin/”” directory of the
binary packages.
.. .. figure:: ../images/anaevensimplerviewer.png
'''
import anatomist.direct.api as ana
from soma import aims
from soma.aims import colormaphints
import sys
import os
from soma.qt_gui import qt_backend
from soma.qt_gui.qt_backend import Qt, loadUi
import six
from six.moves import zip
def findChild(x, y): return Qt.QObject.findChild(x, Qt.QObject, y)
close_qt = False
if 'sphinx_gallery' in sys.modules \
or ('IPython' in sys.modules
and Qt.QApplication.instance() is None):
run_qt = False
close_qt = True
elif Qt.QApplication.instance() is None:
run_qt = True
else:
run_qt = False
# start the Anatomist engine, in batch mode (no main window)
a = ana.Anatomist('-b')
# load the anasimpleviewer GUI
uifile = 'anasimpleviewer-qt4.ui'
anasimpleviewerdir = os.path.join(six.text_type(a.anatomistSharedPath()),
'anasimpleviewer')
cwd = os.getcwd()
# PyQt4 uic doesn' seem to allow specifying the directory when looking for
# icon files: we have no other choice than globally changing the working
# directory
os.chdir(anasimpleviewerdir)
awin = loadUi(os.path.join(anasimpleviewerdir, uifile))
os.chdir(cwd)
# global variables: lists of windows, objects, a fusion2d with a number of
# volumes in it
awindows = []
aobjects = []
fusion2d = []
# vieww: parent block widget for anatomist windows
vieww = findChild(awin, 'windows')
viewgridlay = Qt.QGridLayout(vieww)
nviews = 0
# This class holds methods for menu/actions callbacks, and utility functions
# like load/view objects, remove/delete, etc.
class AnaSimpleViewer(Qt.QObject):
def __init__(self):
Qt.QObject.__init__(self)
self.filedialogdir = '.'
def createWindow(self, wintype='Axial'):
'''Opens a new window in the windows grid layout.
The new window will be set in MNI referential, and have no
menu/toolbars.
'''
global vieww, nviews
x = nviews % 2
y = nviews // 2
nviews += 1
w = a.createWindow(wintype, no_decoration=True, options={'hidden': 1})
w.setAcceptDrops(False)
viewgridlay.addWidget(w.getInternalRep(), x, y)
# keep it in anasimpleviewer list of windows
awindows.append(w)
a.assignReferential(a.mniTemplateRef, w)
# force redrawing in MNI orientation
# (there should be a better way to do so...)
if wintype == 'Axial':
w.muteAxial()
elif wintype == 'Coronal':
w.muteCoronal()
elif wintype == 'Sagittal':
w.muteSagittal()
elif wintype == 'Oblique':
w.muteOblique()
# set a black background
a.execute('WindowConfig', windows=[w],
light={'background': [0., 0., 0., 1.]})
def loadObject(self, fname):
'''Load an object and display it in all anasimpleviewer windows
'''
obj = a.loadObject(fname)
self.registerObject(obj)
def registerObject(self, obj):
'''Register an object in anasimpleviewer objects list, and display it
'''
findChild(awin, 'objectslist').addItem(obj.name)
# keep it in the global list
aobjects.append(obj)
if obj.objectType == 'VOLUME':
# volume are checked for possible adequate colormaps
hints = colormaphints.checkVolume(
ana.cpp.AObjectConverter.aims(obj))
obj.attributed()['colormaphints'] = hints
bb = obj.boundingbox()
# create the 4 windows if they don't exist
if len(awindows) == 0:
self.createWindow('Coronal')
self.createWindow('Axial')
self.createWindow('Sagittal')
self.createWindow('3D')
# set a cool angle of view for 3D
a.execute('Camera', windows=[awindows[-1]],
view_quaternion=[0.404603, 0.143829, 0.316813, 0.845718])
# view obj in these views
self.addObject(obj)
# set the cursot at the center of the object (actually, overcome a bug
# in anatomist...)
position = (aims.Point3df(bb[1][:3]) - bb[0][:3]) / 2.
t = a.getTransformation(obj.getReferential(),
awindows[0].getReferential())
if t:
position = t.transform(position)
a.execute('LinkedCursor', window=awindows[0], position=position)
def addVolume(self, obj, opts={}):
'''Display a volume in all windows.
If several volumes are displayed, a Fusion2D will be built to wrap all
of them.
'''
global fusion2d
if obj in fusion2d:
return
if len(fusion2d) == 0:
# only one object
fusion2d = [None, obj]
else:
# several objects: fusion them
fusobjs = fusion2d[1:] + [obj]
f2d = a.fusionObjects(fusobjs, method='Fusion2DMethod')
if fusion2d[0] is not None:
# destroy the previous fusion
a.deleteObjects(fusion2d[0])
else:
a.removeObjects(fusion2d[1], awindows)
fusion2d = [f2d] + fusobjs
# repalette( fusobjs )
obj = f2d
if obj.objectType == 'VOLUME':
# choose a good colormap for a single volume
if 'volume_contents_likelihoods' in \
obj.attributed()['colormaphints']:
cmap = colormaphints.chooseColormaps(
(obj.attributed()['colormaphints'], ))
obj.setPalette(cmap[0])
else:
# choose good colormaps for the current set of volumes
hints = [x.attributed()['colormaphints'] for x in obj.children]
children = [x for x, y in zip(obj.children, hints)
if 'volume_contents_likelihoods' in y]
hints = [x for x in hints
if 'volume_contents_likelihoods' in x]
cmaps = colormaphints.chooseColormaps(hints)
for x, y in zip(children, cmaps):
x.setPalette(y)
a.addObjects(obj, awindows, **opts)
def removeVolume(self, obj, opts={}):
'''Hides a volume from views (low-level function: use removeObject)
'''
global fusion2d
if obj in fusion2d:
fusobjs = [o for o in fusion2d[1:] if o != obj]
if len(fusobjs) >= 2:
f2d = a.fusionObjects(fusobjs, method='Fusion2DMethod')
else:
f2d = None
if fusion2d[0] is not None:
a.deleteObjects(fusion2d[0])
else:
a.removeObjects(fusion2d[1], awindows)
if len(fusobjs) == 0:
fusion2d = []
else:
fusion2d = [f2d] + fusobjs
# repalette( fusobjs )
if f2d:
obj = f2d
elif len(fusobjs) == 1:
obj = fusobjs[0]
else:
return
a.addObjects(obj, awindows, **opts)
def addObject(self, obj):
'''Display an object in all windows
'''
opts = {}
if obj.objectType == 'VOLUME':
# volumes have a specific function since several volumes have to be
# fusionned, and a volume rendering may occur
self.addVolume(obj, opts)
return
elif obj.objectType == 'GRAPH':
opts['add_graph_nodes'] = 1
a.addObjects(obj, awindows, **opts)
def removeObject(self, obj):
'''Hides an object from views
'''
if obj.objectType == 'VOLUME':
self.removeVolume(obj)
else:
a.removeObjects(obj, awindows, remove_children=True)
def fileOpen(self):
'''File browser + load object(s)
'''
fdialog = Qt.QFileDialog()
fdialog.setDirectory(self.filedialogdir)
fdialog.setFileMode(fdialog.ExistingFiles)
res = fdialog.exec_()
if res:
fnames = fdialog.selectedFiles()
self.filedialogdir = fdialog.directory()
for fname in fnames:
self.loadObject(six.text_type(fname))
def selectedObjects(self):
'''list of objects selected in the list box on the upper left panel
'''
olist = findChild(awin, 'objectslist')
sobjs = []
for o in olist.selectedItems():
sobjs.append(six.text_type(o.text()).strip('\0'))
return [o for o in aobjects if o.name in sobjs]
def editAdd(self):
'''Display selected objects'''
objs = self.selectedObjects()
for o in objs:
self.addObject(o)
def editRemove(self):
'''Hide selected objects'''
objs = self.selectedObjects()
for o in objs:
self.removeObject(o)
def editDelete(self):
'''Delete selected objects'''
objs = self.selectedObjects()
for o in objs:
self.removeObject(o)
olist = findChild(awin, 'objectslist')
for o in objs:
olist.takeItem(olist.row(olist.findItems(o.name,
Qt.Qt.MatchExactly)[0]))
global aobjects
aobjects = [o for o in aobjects if o not in objs]
a.deleteObjects(objs)
def closeAll(self):
'''Exit'''
print("Exiting")
global vieww, awindows, fusion2d, aobjects, anasimple
global awin, viewgridlay
# remove windows from their parent to prevent them to be brutally
# deleted by Qt.
w = None # ensure there is a w variable before the "del" later
for w in awindows:
w.hide()
viewgridlay.removeWidget(w.internalRep._get())
w.setParent(None)
del w
del viewgridlay, vieww
del anasimple
del awindows, fusion2d, aobjects
awin.close()
del awin
a = ana.Anatomist()
a.close()
# instantiate the machine
anasimple = AnaSimpleViewer()
# connect GUI actions callbacks
findChild(awin, 'fileOpenAction').triggered.connect(anasimple.fileOpen)
findChild(awin, 'fileExitAction').triggered.connect(anasimple.closeAll)
findChild(awin, 'editAddAction').triggered.connect(anasimple.editAdd)
findChild(awin, 'editRemoveAction').triggered.connect(anasimple.editRemove)
findChild(awin, 'editDeleteAction').triggered.connect(anasimple.editDelete)
# display on the whole screen
awin.showMaximized()
# tweak: override some user config options
a.config()['setAutomaticReferential'] = 1
a.config()['windowSizeFactor'] = 1.
# run Qt
if __name__ == '__main__':
if run_qt:
Qt.QApplication.instance().exec_()
elif 'sphinx_gallery' in sys.modules:
# load a data
awin.showNormal()
awin.resize(1000, 700) # control the size of the snapshot
anasimple.loadObject('irm.ima')
# these 2 events ensure things are actually drawn
Qt.QApplication.instance().processEvents()
Qt.QApplication.instance().processEvents()
# snapshot the whole widget
if Qt.QT_VERSION >= 0x050000:
ws = awin.grab() # Qt5 only
else:
ws = Qt.QPixmap.grabWidget(awin) # Qt4 only
# openGl areas are not rendered in the snapshot, we have to make them
# by hand
p = Qt.QPainter(ws)
for w in awindows:
s = w.snapshotImage()
# draw the GL rendering into the image (pixmap)
p.drawImage(w.mapTo(awin, Qt.QPoint(0, 0)), s)
del w, s, p
# snapshot to matplotlib
import matplotlib
# backend = matplotlib.get_backend()
matplotlib.use('agg', force=True) # force agg
from matplotlib import pyplot
im = qt_backend.qimage_to_np(ws)
plot = pyplot.imshow(im)
axes = pyplot.gcf().axes
axes[0].get_xaxis().set_visible(False)
axes[0].get_yaxis().set_visible(False)
pyplot.show(block=False)
# matplotlib.use(backend, force=True) # restore backend
if close_qt:
anasimple.closeAll()
Using PyAnatomist in BrainVisa¶
This example shows how to use Anatomist from a BrainVisa process, and get events from Anatomist even when it is run through a socket connection.
Download source: bvProcessSocket.py
# -*- coding: utf-8 -*-
# This software and supporting documentation are distributed by
# Institut Federatif de Recherche 49
# CEA/NeuroSpin, Batiment 145,
# 91191 Gif-sur-Yvette cedex
# France
#
# This software is governed by the CeCILL license version 2 under
# French law and abiding by the rules of distribution of free software.
# You can use, modify and/or redistribute the software under the
# terms of the CeCILL license version 2 as circulated by CEA, CNRS
# and INRIA at the following URL "http://www.cecill.info".
#
# As a counterpart to the access to the source code and rights to copy,
# modify and redistribute granted by the license, users are provided only
# with a limited warranty and the software's author, the holder of the
# economic rights, and the successive licensors have only limited
# liability.
#
# In this respect, the user's attention is drawn to the risks associated
# with loading, using, modifying and/or developing or reproducing the
# software by the user in light of its specific status of free software,
# that may mean that it is complicated to manipulate, and that also
# therefore means that it is reserved for developers and experienced
# professionals having in-depth computer knowledge. Users are therefore
# encouraged to load and test the software's suitability as regards their
# requirements in conditions enabling the security of their systems and/or
# data to be ensured and, more generally, to use and operate it in the
# same conditions as regards security.
#
# The fact that you are presently reading this means that you have had
# knowledge of the CeCILL license version 2 and that you accept its terms.
'''
Using PyAnatomist in BrainVisa
------------------------------
This example shows how to use Anatomist from a BrainVisa process, and get events from Anatomist even when it is run through a socket connection.
'''
from __future__ import print_function
from __future__ import absolute_import
from neuroProcesses import *
from brainvisa.tools import aimsGlobals
import string
name = 'Test anatomist API socket implementation'
userLevel = 0
# by default, socket implementation is loaded in brainvisa
from brainvisa import anatomist as pyanatomist
import anatomist.api
signature = Signature(
'object_to_load', ReadDiskItem(
"3D Volume", aimsGlobals.anatomistVolumeFormats),
)
def initialization(self):
pass
def execution(self, context):
# with attribute create is False, the constructor returns the existing
# instance of anatomist or None if there isn't one.
a = pyanatomist.Anatomist(create=False)
print("anatomist instance:", a)
a = pyanatomist.Anatomist(create=True)
print("anatomist instance:", a)
# register a function to be called when an object is loaded in anatomist
a.onLoadNotifier.add(display)
# a.onLoadNotifier.remove(display)
# a.onCreateWindowNotifier.add(display)
# a.onDeleteNotifier.add(display)
# a.onFusionNotifier.add(display)
# a.onCloseWindowNotifier.add(display)
# a.onAddObjectNotifier.add(display)
# a.onRemoveObjectNotifier.add(display)
# a.onCursorNotifier.add(display)
obj = a.loadObject(self.object_to_load)
w = a.createWindow('Axial')
a.addObjects([obj], [w])
ref = obj.referential
context.write("referential of object :", ref.refUuid)
return [obj, w, ref]
def display(event, params):
print("** Event ** ", event, params)