'This pipeline helps you to choose the best reconstruction for a PET camera by quantifying activity in jaszczak's spheres.
Before using this pipeline:
All this steps can be openned separatly, using right click.
- Define your jaszczak if it is a new one.
- Import your CT and PET Dicom images
- Convert your dicom images into nii format.
Finally, you can use this pipeline by selecting just your CT image (others parameters are automatically deducted).
' The Jaszczak SPECT Phantom provides consistent performance information for any SPECT or PET system.'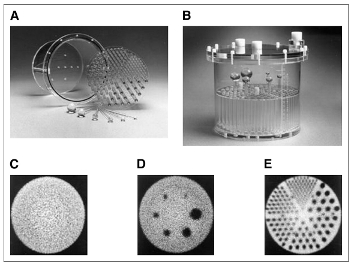
D : radioactivity is injected in spheres called "hot spheres", the others are "cold" ( you can see these ones in black). Then the activity in spheres' image is quantified and compare with the theoretical one, for each reconstruction. A graph allows you to compare reconstructions E : Rods are used to check visually the image resolution. To do so, all slices with rods are summed. To quantify spheres:
To quantify the activity in Jaszczak's spheres, you need to fill the fields about the activity injected in Jaszczak and its spheres.quantify spheres.
Pipeline steps' details :
The acquisition can be dynamic, so the mean is computed (step 1) All PET images and the CT one are segmented to check a possible gap between them. (step 2 Segment sph PET , 3 Segment sph CT, 4 Check fusion PET CT) Note : If so, the camera should be fixed. According to this gap, the spheres' mask is computed (step 5 Build Sph Mask) and used to quantify spheres (step 7 Quantify spheres) All slices with rods are summed and then displayed, to check visually the resolution (step 6 Sum rods) All reconstructions are quantified (step 7 Quantify spheres), using ROI previously computed and AimsRoiFeatures All reconstructions can be compared thanks to the graphic computed in step 8 Compare quantification
Pipeline steps' documentation :
Spheres Segmentations :
Display CT, PET, ROI and Mask fusions
Reconstruction Comparison :
'
database: Choice ( input )
display_separatly_each_process: Choice ( input )
Pets_mean: ListOf( Phantom PET Mean ) ( input )
CT: Phantom CT ( input )
phantom: String ( input )
center: String ( input )
jzkSpec: Phantom Spec ( input )
theoric_values: Phantom Theoric Values ( optional, output )
bkg_preparedActivity: Float ( input )prepared activity in background unit is Mega bequerel
bkg_H_M_pre: String ( input )
bkg_residualActivity: Float ( input )residual activity in background unit is Mega bequerel
bkg_H_M_res: String ( input )
sph_preparedActivity: Float ( input )prepared activity in spheres unit is Mega bequerel
sph_H_M_pre: String ( input )
sph_residualActivity: Float ( input )residual activity in spheres unit is Mega bequerel
sph_H_M_res: String ( input )
H_M_acq: String ( input )
periode_isotope: Float ( input )
run_parallel: Boolean ( input )
verbose: Boolean ( input )
reconstructionNames: ListOf( String ) ( input )
Pets_segmented: ListOf( Phantom PET Mean Segmented ) ( output )
ct_segmented: Phantom CT Segmented ( output )
sph_rois: Phantom ROI Sph ( optional, output )
sph_roi_in_Noise: Phantom ROI Sph inNoise ( output )
PET_mean_toDisplay: Phantom PET Mean ( output )
Pet_segmented_toDisplay: Phantom PET Mean Segmented ( output )
CT2PetMeans: ListOf( CT to PET transformation file ) ( output )
PETs_ghosts: ListOf( Phantom ROI Sph Ghosts ) ( output )
allRods: ListOf( Phantom Rods ) ( output )
allRods_sum: ListOf( Phantom Rods Sum ) ( output )
stats_inSpheres: ListOf( Phantom Activity Stats ) ( output )
stats_inNoise: ListOf( Phantom Noise Stats ) ( output )
stats_inGhosts: ListOf( Phantom ROI Sph Ghosts Stat ) ( output )
ratios_inSpheres: ListOf( Phantom Result ) ( output )
ratios_inGhosts: ListOf( Phantom Sph Ghosts Result ) ( output )
graph: Phantom Graph HotSph ( output )
graph_coldSph: Phantom Graph ColdSph ( output )
Spheres_ID_PVC: String ( input )
Spheres_ID_FWHM: String ( optional, input )
selected_FWHM_prieto: String ( optional, input )
selected_FWHM_normal: String ( optional, input )
selected_FWHM_special: String ( optional, input )
ROI_folders_list: ListOf( Phantom ROI PVC ) ( optional, output )
FWHM_informations_files_list: ListOf( FWHM Informations Jaszczak ) ( optional, output )
Chi_curves_folders_list: ListOf( Chi Square folder ) ( optional, output )
Corrected_uptakes_list: ListOf( Phantom PVC Corrected Uptakes Stats ) ( optional, output )
RC_calculation_method: Choice ( optional, input )
PVC_method: Choice ( optional, input )
Fill_centers: Choice ( optional, input )
CT_sph1_center: Point3D ( optional, input )
CT_sph2_center: Point3D ( optional, input )
CT_sph3_center: Point3D ( optional, input )
CT_sph4_center: Point3D ( optional, input )
CT_sph5_center: Point3D ( optional, input )
CT_sph6_center: Point3D ( optional, input )
PVC_dictionary: PVC Jaszczak dictionary ( optional, output )
Toolbox : Nuclear Imaging
User level : 0
Identifier :
JZKPipelineFile name :
brainvisa/toolboxes/nuclearimaging/processes/JZKPipeline.pySupported file formats :
Pets_mean :NIFTI-1 image, NIFTI-1 imageCT :NIFTI-1 image, NIFTI-1 imagejzkSpec :CSV file, CSV filetheoric_values :CSV file, CSV filePets_segmented :NIFTI-1 image, NIFTI-1 imagect_segmented :NIFTI-1 image, NIFTI-1 imagesph_rois :NIFTI-1 image, NIFTI-1 imagesph_roi_in_Noise :NIFTI-1 image, NIFTI-1 imagePET_mean_toDisplay :NIFTI-1 image, NIFTI-1 imagePet_segmented_toDisplay :NIFTI-1 image, NIFTI-1 imageCT2PetMeans :Transformation matrix, Transformation matrixPETs_ghosts :NIFTI-1 image, NIFTI-1 imageallRods :NIFTI-1 image, NIFTI-1 imageallRods_sum :NIFTI-1 image, NIFTI-1 imagestats_inSpheres :CSV file, CSV filestats_inNoise :CSV file, CSV filestats_inGhosts :CSV file, CSV fileratios_inSpheres :CSV file, CSV fileratios_inGhosts :CSV file, CSV filegraph :CSV file, CSV filegraph_coldSph :CSV file, CSV fileROI_folders_list :Directory, DirectoryFWHM_informations_files_list :Text file, Text fileChi_curves_folders_list :Directory, DirectoryCorrected_uptakes_list :CSV file, CSV filePVC_dictionary :Text file, Text file