


Compute a mesh of one or two hemispheres
This process create a mesh of each cerebral hemisphere. It include a pretreatment, which open the sulcus.
This new version yields a spherical mesh, which allows the inflation of the mesh as the grey/white mesh.
Side: Choice ( input )Left, Right or Both Hemispheres
mri_corrected: T1 MRI Bias Corrected ( input )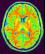
split_mask: Split Brain Mask ( input )
left_hemi_cortex: Left CSF+GREY Mask ( input )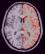
right_hemi_cortex: Right CSF+GREY Mask ( input )
left_hemi_mesh: Left Hemisphere Mesh ( output )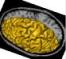
right_hemi_mesh: Right Hemisphere Mesh ( output )
fix_random_seed: Boolean ( input )
Toolbox : Morphologist
User level : 2
Identifier :
GetSphericalHemiSurfaceSupported file formats :
mri_corrected :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_mask :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imageleft_hemi_cortex :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imageright_hemi_cortex :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 imageleft_hemi_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI meshright_hemi_mesh :GIFTI file, GIFTI file, MESH mesh, MNI OBJ mesh, PLY mesh, TRI mesh