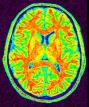

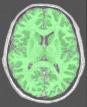



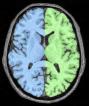
Split the brain into three parts (hemispheres + cerebellum)
This procedure aims at splitting the two hemispheres and at removing the cerebellum and a part of brain stem in order to give access to the internal and low faces of the cortex.
An erosion is applied to a mask of white matter in order to split it at the levels of corpus callosum and pons. This operation provides 3 seeds corresponding to the two hemispheres and cerebellum. Then, these seeds grow first inside white matter and finally throughout grey matter in order to recover the hemisphere shapes.
brain_mask: T1 Brain Mask ( input )
t1mri_nobias: T1 MRI Bias Corrected ( input )
histo_analysis: Histo Analysis ( input )
commissure_coordinates: Commissure coordinates ( optional, input )
use_ridges: Boolean ( input )
white_ridges: T1 MRI White Matter Ridges ( input )
use_template: Boolean ( input )
split_template: Hemispheres Template ( input )
mode: Choice ( input )
variant: Choice ( input )
bary_factor: Choice ( input )between 0 and 1.
mult_factor: Choice ( optional, input )
initial_erosion: Float ( input )
cc_min_size: Integer ( input )
split_brain: Split Brain Mask ( output )
fix_random_seed: Boolean ( input )
Toolbox : Morphologist
User level : 0
Identifier :
SplitBrainFile name :
brainvisa/toolboxes/morphologist/processes/segmentationpipeline/components/SplitBrain.pySupported file formats :
brain_mask :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imaget1mri_nobias :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysiscommissure_coordinates :Commissure coordinates, Commissure coordinateswhite_ridges :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_template :gz compressed NIFTI-1 image, Aperio svs, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, Ventana bif, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_brain :gz compressed NIFTI-1 image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIS image, JPEG image, MINC image, NIFTI-1 image, SPM image, TIFF image, TIFF(.tif) image, gz compressed MINC image, gz compressed NIFTI-1 image