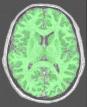
Compute a binary mask of the brain from a bias corrected T1-weighted image
The basic principles of this procedure are described in Ana Brain Mask from T1 MRI and Correction Brain Mask from T1 MRI. Moreover, the log files are full of information...
In this low level access, you can tune some parameters to overcome difficult cases. If it is still a failure, your last hope is in using the commandline procedure, which gives access to much more parameters. An interesting alternative, however, is manual drawing, at least to clean up problematic areas... Semi-manual drawing may be especially wellcome when a lesion is disturbing the segmentation process.
For more information:
Robust brain segmentation using histogram scale-space analysis and mathematical morphology, J.-F. Mangin, O. Coulon, and V. Frouin MICCAI, MIT, LNCS-1496, Springer Verlag 1230-1241, 1998
You can also try the commandline
VipGetBrain -help
mri_corrected: T1 MRI Bias Corrected ( input )
histo_analysis: Histo Analysis ( input )
Grey/white statistics
mode: Choice ( input )
Commissure_coordinates: Commissure coordinates ( optional, input )
brain_mask: T1 Brain Mask ( output )
binary mask
regularization: Boolean ( input )during binarization
erosion_size: Float ( input )to get brain seed
layer: Choice ( input )
first_slice: Integer ( input )slices to erase at top
last_slice: Integer ( input )slices to erase at bottom
lesion_mask: Lesion Mask ( optional, input )binary mask (option)
Toolbox : Morphologist
User level : 2
Identifier :
VipGetBrainSupported file formats :
mri_corrected :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo AnalysisCommissure_coordinates :Commissure coordinates, Commissure coordinatesbrain_mask :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagelesion_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 image