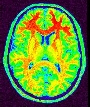



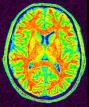

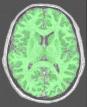
Compute a binary mask of the brain from a raw MR scan
This procedure relies on the fact that the brain is a light and massive object surrounded by a black strip related to the skull, which largely isolates it from the other tissues. The image analysis procedure aims at splitting the few connexions between the brain and the external world using an erosion process very similar to geological phenomena or to the action of the rust, when it weakens metallic objects at the level of their bottlenecks before breaking them.
The grey level image is thresholded first in order to get a binary object (white on black background). During a second stage, an erosion process is simulated in order to trim the object until a given thickness. This erosion fathers several disconnected pieces. The largest one is selected as a seed of the brain. A dilation process makes the seed growth in order to recover the actual brain shape.
An illustration of the process at the level of a slice:
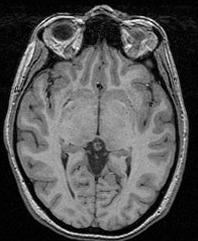
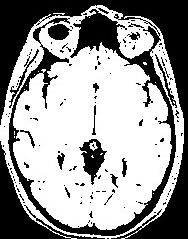
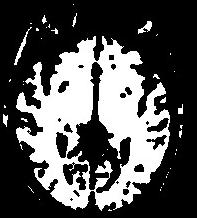

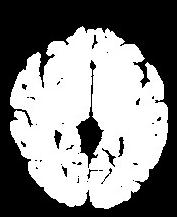
In order to reach its goal, this line puts together 3 steps:The segmentation process embeds a lot of refinements aiming at overcoming some problems occuring with some specific sequences or unusual morphologies. You can have a glipmse of these refinements if you open the log window. The main one is the automatic tuning of the erosion size.
- A correction of the spatial bias (Vip Bias Correction). It is the key step of the whole process. If this correction is not close to perfect, the following results may suffer. Hence, it is important to check the bias correction behaviour on a few datasets from your scanner before going further. The two initial choices (contrast, bias type) are dedicated to this procedure.
- A histogram analysis (Vip Histogram Analysis)
- A brain segmentation (Vip Get Brain)
If the file including the localisation of the commisures has been filled, a final step will try to remove possible residuals related to sinus/meninges at the level of interhemispheric plane.
If a lesion mask has been provided, this area will be added to the initial binarisation whatever the greylevels inside. It will overcome situations where a part of the brain is lost because the erosion process leads to several brain seeds.
Checking result and problem solving:
Cf. brainVISA tools:
Validation Brain Mask from T1 MRI
Correction Brain Mask from T1 MRI
You can for instance visualize simultaneously the bias corrected image and the brain mask, relying onAnatomist linked cursor:
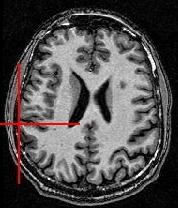
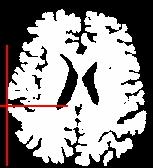
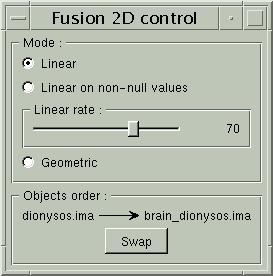
Another way consists of the two volume fusion. brainVISA's eye can do it for you. If you prefer manual work, select the two volumes, give a unicolor colormap with the ufusion extension to the mask, and choose the linear combination fusion mode:
If all is OK, you can go further...
You can signal to brainVISA that this result has to be frozen (not recomputed...) using the procedure:Validation Brain Mask from T1 MRI
Otherwise, you have to execute the error procedure related to this processing line:
Correction Brain Mask from T1 MRI
T1mri: Raw T1 MRI ( input )
Contrast: Choice ( input )
Bias_type: Choice ( input )
mri_corrected: T1 MRI Bias Corrected ( output )
histo_analysis: Histo Analysis ( output )
brain_mask: T1 Brain Mask ( output )
Commissure_coordinates: Commissure coordinates ( optional, input )
lesion_mask: 3D Volume ( optional, input )
Toolbox : Morphologist
User level : 2
Identifier :
AnaT1toBrainMaskSupported file formats :
T1mri :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagemri_corrected :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysisbrain_mask :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageCommissure_coordinates :Commissure coordinates, Commissure coordinateslesion_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 image