Split the brain into three parts (hemispheres + cerebellum)
The basic principle of this procedure is described in Ana Split Brain From Brain Mask. Furthermore, a lot of information may be found in the log file.
For more information and to discover a funny alternative to the mathematical morphology based approach:
Shape bottlenecks and Conservative Flow systems, J.- F. Mangin, J. Regis and V. Frouin IEEE/SIAM MMBIA Workshop (Math. Methods in Biomed. Image Analysis), San Francisco, IEEE Press 319-328, 1996
You can also try the commandline:
VipSplitBrain -help
mri_corrected: T1 MRI Bias Corrected ( input )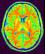
Use_template: Boolean ( input )
split_template: Hemispheres Template ( optional, input )
brain_mask: T1 Brain Mask ( input )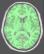
histo_analysis: Histo Analysis ( input )
split_mask: Split Brain Mask ( output )
Commissure_coordinates: Commissure coordinates ( optional, input )
white_algo: Choice ( optional, input )
mult_factor: Float ( optional, input )threshold for white matter: mean - mult_factor*sigma
bary_factor: Float ( input )threshold for white matter as barycenter between grey and white
white_threshold: Integer ( optional, input )direct input
initial_erosion: Float ( input )
cc_min_size: Integer ( input )
Toolbox : Morphologist
User level : 2
Identifier :
VipSplitBrainSupported file formats :
mri_corrected :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_template :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagebrain_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysissplit_mask :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageCommissure_coordinates :Commissure coordinates, Commissure coordinates