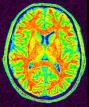

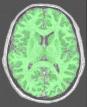




Split the brain mask into three parts: hemispheres + cerebellum/stem (procedure built upon Vip Split Brain)
This procedure aims at splitting the two hemispheres and at removing the cerebellum and a part of brain stem in order to give access to the internal and low faces of the cortex. It is built upon an idea similar to the one used to segment the brain (Brain Mask From T1 MRI). An erosion is applied to a mask of white matter in order to split it at the levels of corpus callosum and pons. This operation provides 3 seeds corresponding to the two hemispheres and cerebellum. Then, these seeds grow first inside white matter and finally throughout grey matter in order to recover the hemisphere shapes.
The resulting image is called a Voronoï diagram, namely the parcelling of a domain related to the distance to the closest seed. This diagram looks like the influence zones of middle-age castles, which role is played by the seeds.
An illustration of this "split and grow" idea applied here with 5 seeds:
A first Voronoï diagram is built from the 5 largest seeds stemming from a white matter erosion:
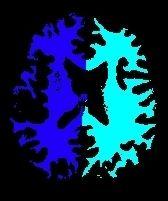
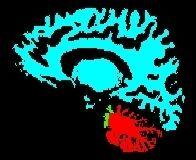
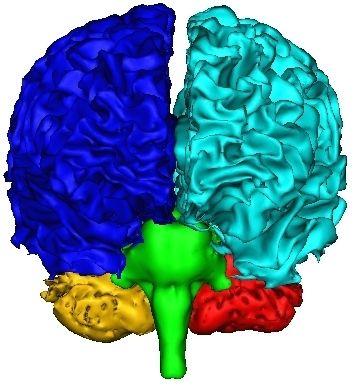
This first diagram yields the seeds for the computation of a second diagram throughout the whole brain:
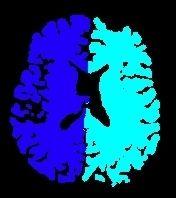

If the commissure localization is provided by the user, some holes will be dug inside corpus callosum and brain stem in order to speed up the split.
If a template of the Voronoï diagram in Talairach space is provided, it will help the procedure to select the erosion size which leads to the best seeds. Thanks to the template, moreover, a given anatomical entity can be represented by a seed made up of several connected components, which increases the process robustness. The erosion process, indeed, can split the two cerebellum hemispheres, or split the temporal lobe from the ipsilateral cortex hemisphere. In such critical cases, if only one connected component is selected as seed, the Voronoï diagram boundaries may be wrong.
When these two options are selected, the main remaining weakness of the process is located at the level of hemisphere/cerebellum interfaces. This boundary, indeed, which delineates two grey matter areas, is difficult to locate using distance criteria, because partial volume often mix grey and white matter in the cerebellum. We think over that problem...Result checking and problem solving:
Cf. BrainVISA tools:
Validation Split Brain from Brain Mask
Correction Split Brain from Brain Mask
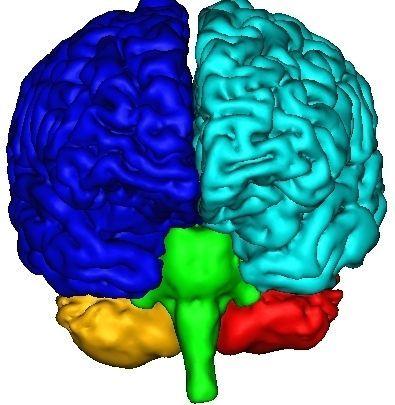
The final result, that you can visualize with a volume fusion obtained from BrainVISA's eye, should look like the following:
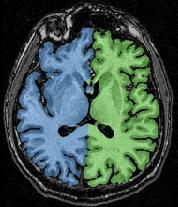
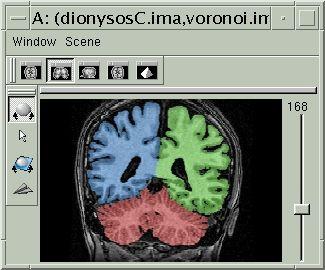

If all seems OK, you can go to the next step...
You can signal to BrainVISA that the result should be frozen using: Validation Split Brain from Brain Mask
Otherwise, you have to execute the associated error procedure (not developped yet, sorry)
Correction Split Brain from Brain Mask
mri_corrected: T1 MRI Bias Corrected ( input )
histo_analysis: Histo Analysis ( input )
brain_mask: T1 Brain Mask ( input )
Use_template: Boolean ( input )
split_template: Hemispheres Template ( optional, input )
split_mask: Split Brain Mask ( output )
Commissure_coordinates: Commissure coordinates ( optional, input )
Toolbox : Morphologist
User level : 2
Identifier :
AnaSplitBrainFromBrainMaskSupported file formats :
mri_corrected :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagehisto_analysis :Histo Analysis, Histo Analysisbrain_mask :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_template :gz compressed NIFTI-1 image, Aperio svs, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, FreesurferMGH, FreesurferMGZ, GIF image, GIS image, Hamamatsu ndpi, Hamamatsu vms, Hamamatsu vmu, JPEG image, Leica scn, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, Sakura svslide, TIFF image, TIFF image, TIFF(.tif) image, TIFF(.tif) image, VIDA image, Ventana bif, XBM image, XPM image, Zeiss czi, gz compressed MINC image, gz compressed NIFTI-1 imagesplit_mask :gz compressed NIFTI-1 image, BMP image, DICOM image, Directory, ECAT i image, ECAT v image, FDF image, GIF image, GIS image, JPEG image, MINC image, NIFTI-1 image, PBM image, PGM image, PNG image, PPM image, SPM image, TIFF image, TIFF(.tif) image, VIDA image, XBM image, XPM image, gz compressed MINC image, gz compressed NIFTI-1 imageCommissure_coordinates :Commissure coordinates, Commissure coordinates