

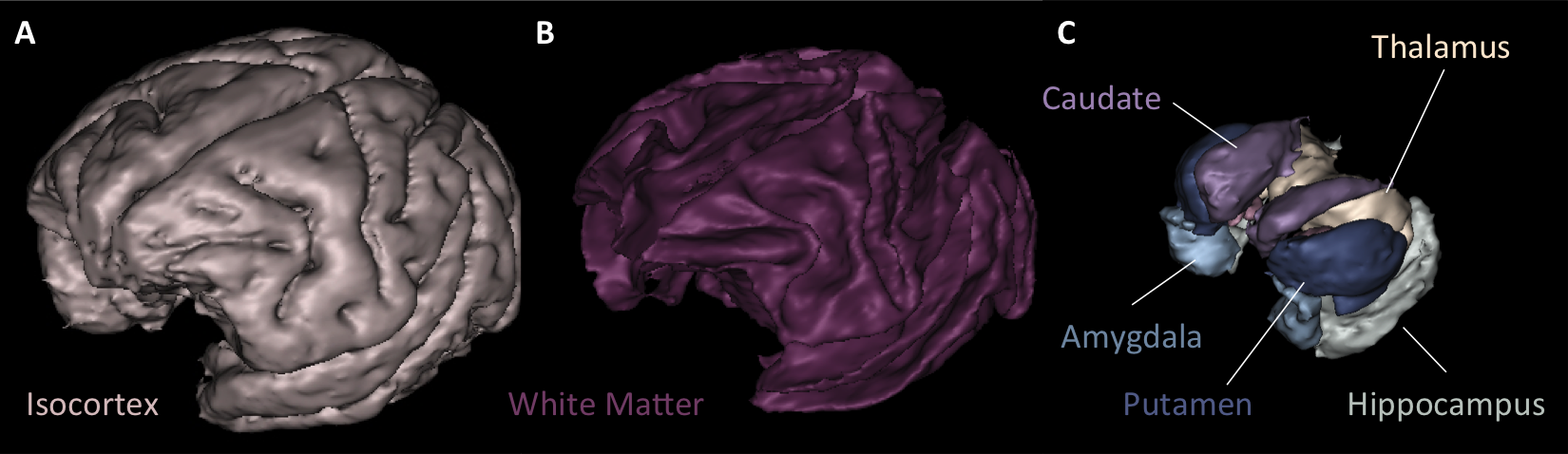
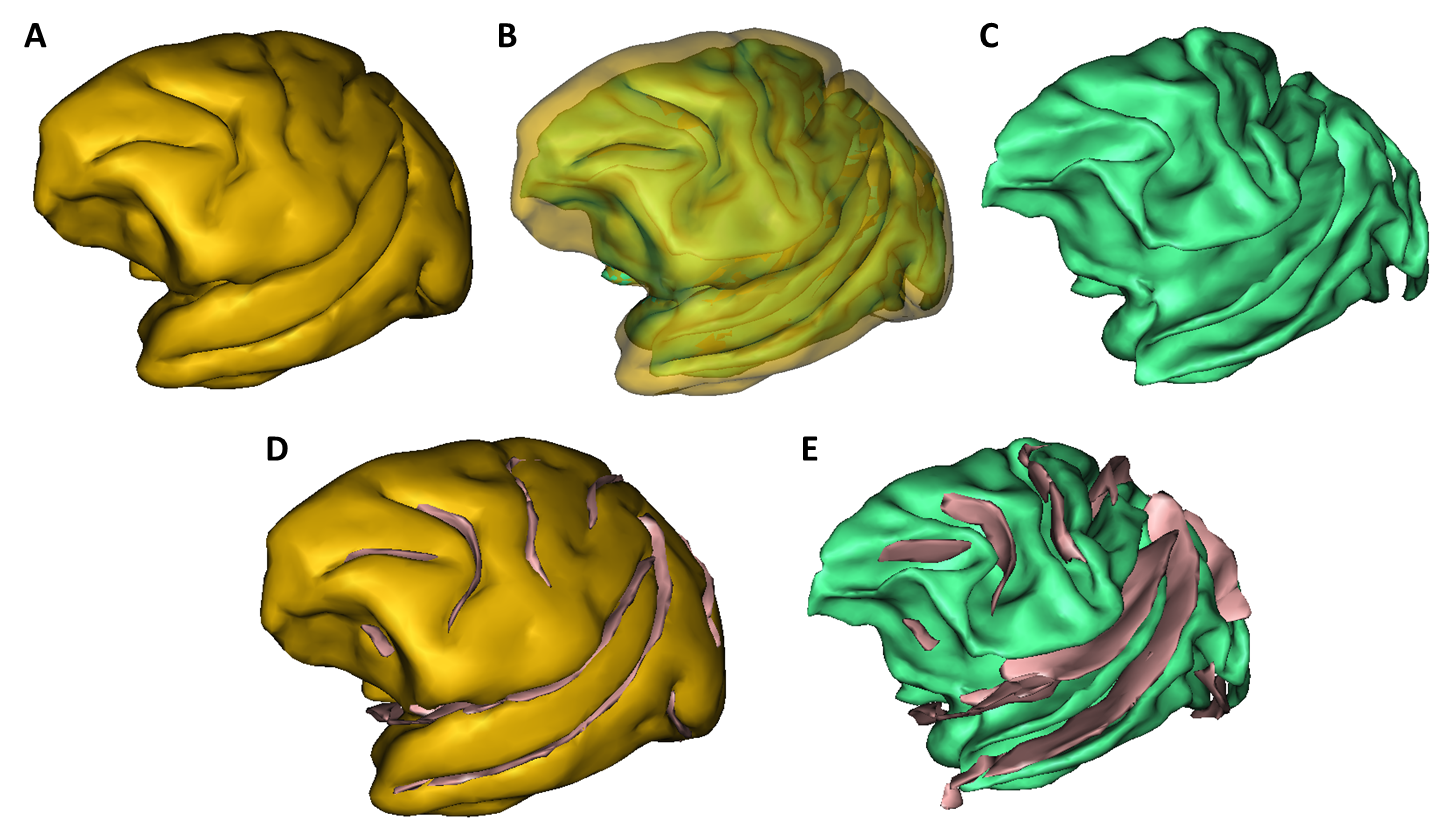
Primatologist is a toolbox dedicated to the analysis of Non-Human Primates (NHPs) in vivo brain images. It is centered around a homonymous pipeline which performs automated parcellations of T1- and T2-weighted MR images of the brain into cortical and subcortical anatomical regions of interest as well as cortical surfaces extraction and sulci segmentation. It depends on the Morphologist toolbox.
Our segmentation method is based on the fitting of a Gaussian Mixture Model (GMM) stemming from a Markov Random Field (MRF) by Expectation-Maximization. An anatomical atlas is first registered towards the target image and "probabilized" to serve as prior probabilities in the GMM scheme. Additionnaly, MRF clique priors were computed from the anatomical atlas and used in the MRF, thus minimizing parameter tuning. Denoising and bias correction are also integrated to this iterative framework.
Emphasis was placed on the robustness of the pipeline. Several preprocessing steps insure minimum failures: a first non-probabilistic bias estimation is performed, followed by a skull-stripping step dedicated to NHPs, whose massive head muscles impair ordinary skull-stripping methods. Good care was also given to the initialization of GMM parameters.
If our method could, in theory, be applied to any NHP, only the Macaque atlas is currently available, limiting in practice its use to this specie.
Tools allowing operations on atlases and hierarchies, volumetry measures and atlas-based analyses of PET data are also embedded.
Do you know how BrainVISA works ? You should start by getting in touch with its philosophy:
The toolbox is made of pipelines that allow the complete processing of input data (import, segmentation) and folders containing individual processes that can be building blocks, analysis tools, viewers, etc. Almost everything should be doable using only two processes: Import MRI for importing your data in a BrainVISA database, and Primatologist for processing them.
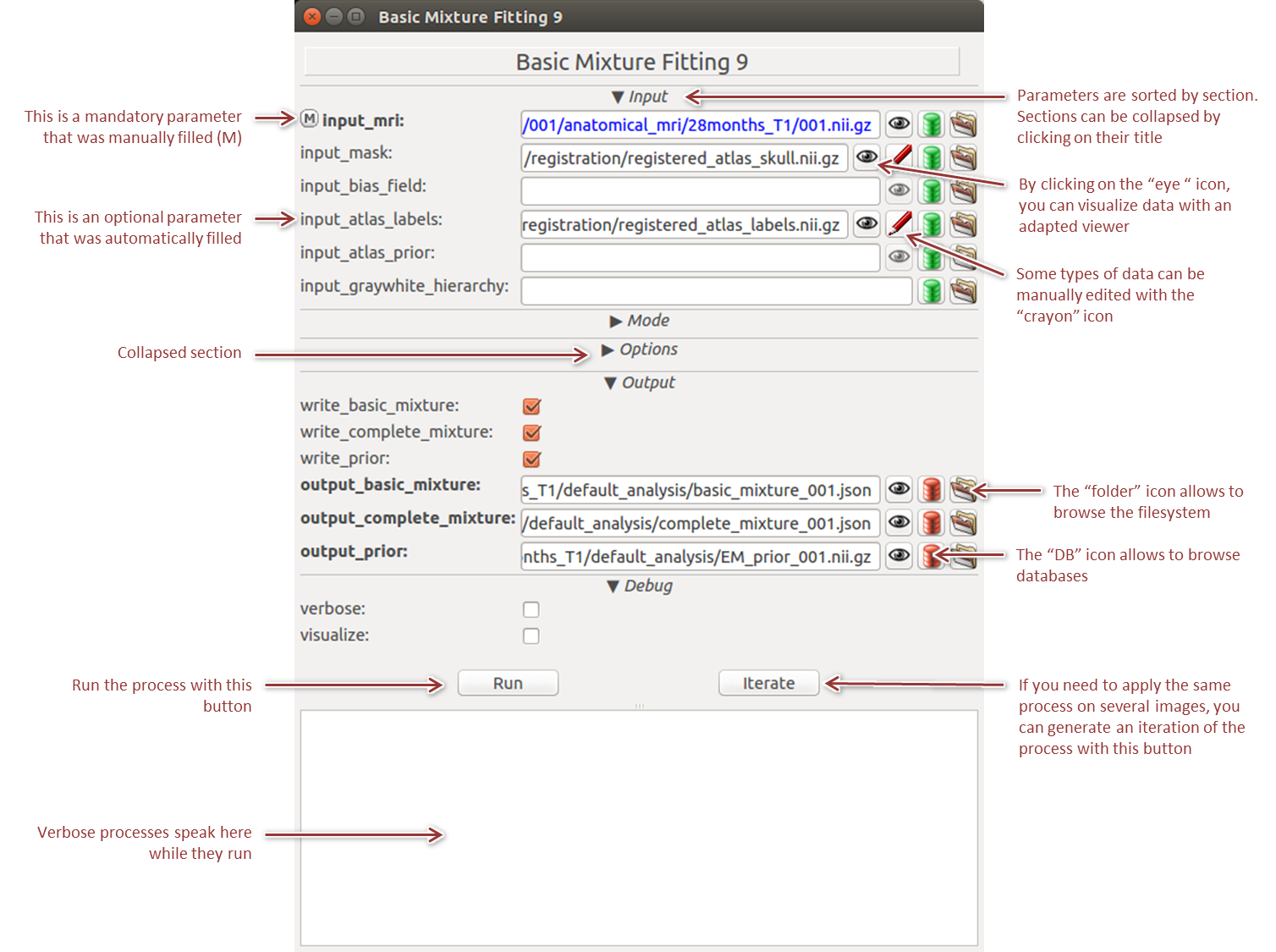
BrainVISA main building blocks are processes, i.e. individual processing units. A process window is composed of a list of inputs and parameters that the user must fill, a few quick links to tools (browse for files, data viewers and editors, etc.) and a context space where the process can communicate when it runs (Figure 3).
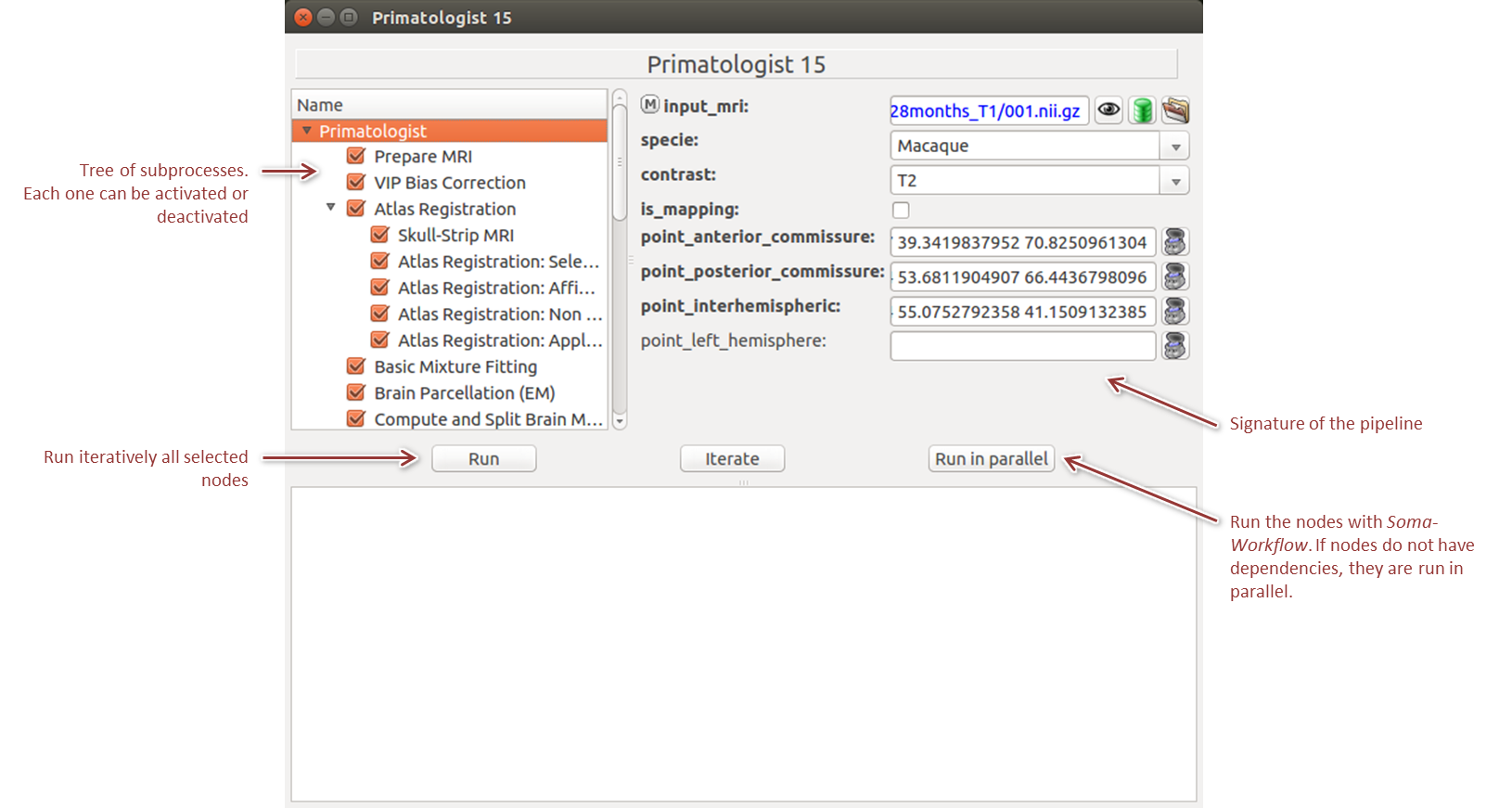
A pipeline is a particular kind of process which consists in the successive or parallel application of several more basic processes. Additionnaly to its signature of parameters (like a process), a pipeline window is composed of a subprocess tree that can be activated or deactivated (Figure 4). It also possesses an additional run button, Run in parallel, which allows to transfer the execution to Soma Workflow, a distributed processing software.
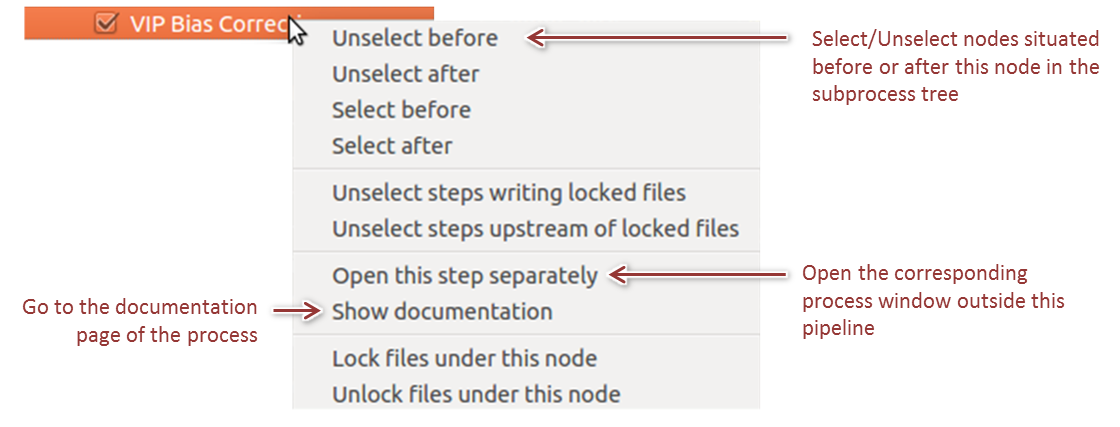
By right clicking on a node in the tree, a contextual menu allows to select/unselect nodes in extensive ways and to access process specific informations (Figure 5).
Primatologist works with BrainVISA's database/ontology system. Everything is explained in the documentation of the importation process.
In order to test this pipeline, we have made available a cross-sectional dataset of 10 T2-weighted MRIs acquired in
Its use is free for academic work upon citation of the following paper:
Primatologist stems from a collaboration between the image processing and analysis teams of MIRCen, led by Thierry Delzescaux, and NeuroSpin, led by Jean-François Mangin. It was primarily developped by Yaël Balbastre during is PhD at MIRCen, a CEA facility dedicated to preclinical imaging, within the Neurodegenerative Diseases Laboratory (UMR n°9199, CEA, CNRS, Université Paris-Saclay).
Primatologist and BrainRAT core maintainers are the same (Nicolas Souedet, Anne-Sophie Hérard, Thierry Delzescaux) and can be contacted by e-mail at brainrat@cea.fr.
Primatologist is distributed under the CeCILL v2.1 license, which is GPL-compatible. Its use is free for academic work. However, in that case, please reference the following paper in your work:
The atlas used in our pipeline was built and published by Evan Calabrese and colleagues at Duke Center for In Vivo Microscopy NIH/NIBIB (P41 EB015897):
If you use results that depend on the Surface and Sulci Extraction pipeline, please also cite:
Methodology and implementation are detailed in the Primatologist article and in Yaël's thesis: