

CONnectivity-based STructural parcELLATION
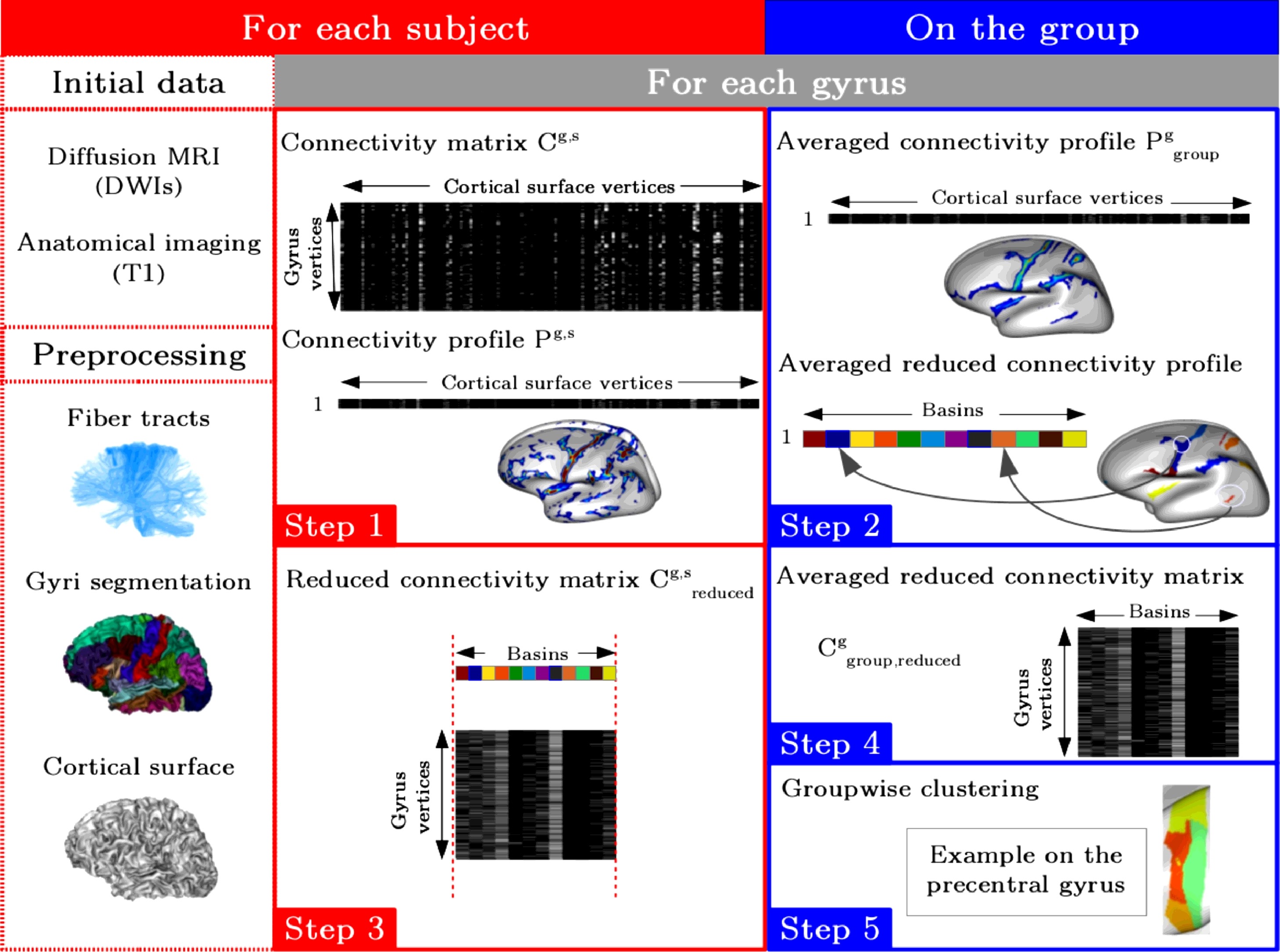
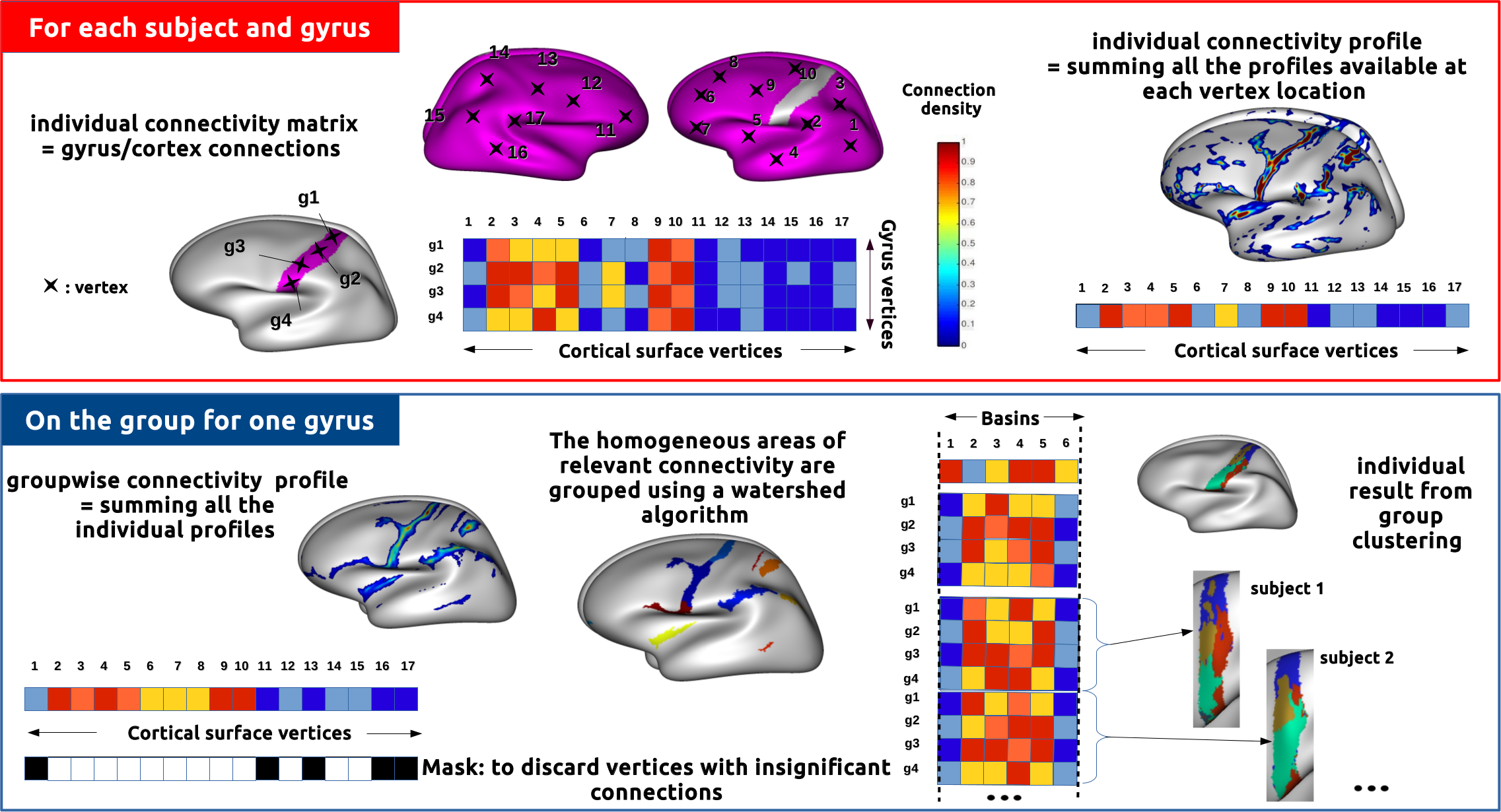
The toolbox Constellation proposes a connectivity-based parcellation framework for the whole cortical surface. The approach chosen performs gyrus by gyrus to parcellate the whole human cortex. The main originality of the method is to compress for each gyrus the connectivity profiles used for the clustering without any anatomical prior information. This step takes into account the interindividual cortical and connectivity variability. To this end, we consider intersubject high density connectivity areas extracted using a surface-based watershed algorithm.
Methods are organized as follow:
There are basically two diffent ways to use Constellation:
Constellation is a somewhat complex toolbox, as it is using different types of data (namely diffusion MRI and anatomical MRI), each preprocessed in potentially different ways by different software. Moreover Constellation includes both intra and inter-subject processings which are at some point intricated. A few choices have to be done by the user.
For this reason, users are advised to read the present documentation before proceeding.
Constellation does not handle by itself the use of thirdparty software to process T1 and diffusion MRI data. The user has thus to take care of running Freesurfer on the T1 MRI (either directly or via the BrainVisa toolbox for Freesurfer), and to process the diffusion MRI and run the fiber tracking or connectivity algorithms (using Connectomist or FSL).
Currently T1 MRI have to be processed using FreeSurfer to take advantage of its surface-based inter-subject correspondance. Freesurfer can be run outside of BrainVISA, or using the BrainVISA / Freesurfer toolbox.
Once Freesurfer has run, there are two final passes that should be run on its outputs. This step features an important choice: the Cortical Grey/white interface mesh resolution and method (see below).
Make meshes of the grey/white interfaces with vertex-vertex correspondance between subjects, and corresponding gyri textures for these meshes. The simplest way to do so is to use the BrainVisa Freesurfer toolbox and its Freesurfer outputs To BrainVisa conversion pipeline.
The toolbox will resample on a template spherical mesh, which is not necessarily Freesurfer's one, so according to the chosen template mesh, it can end up with a different inter-subject correspondance (and a different number of vertices in meshes) than the mri_surf2surf tool from FS.
The pipeline will also produce concatenated, whole brain (both hemispheres) meshes and textures for use with Constellation.
These operations could also partly be done using Freesurfer mris_convert and mri_surf2surf commands to produce GIFTI files and resampled meshes but there would still be coordinates systems issues.
Meshes must be resampled to a common resolution which provides vertex-by-vertex correspondance between subjects. To do so we use the spheric interpolation and the topology of an icosahedron-based template mesh. This can be done either using BrainVisa/Freesurfer toolbox which provides its own template icosphere mesh and interpolation method, or using FreeSurfer, which allows to resample to several icospheres with different resolutions. The choice of the template mesh is very important, because it will determine the corresponance between subjects in the group to be processed. But more importantly, using an atlas to parcellate a new subject will need to use the same template mesh as the atlas.
Constellation has been tested using meshed of 30000-40000 vertices per hemisphere. Using lower resolutions will make results more imprecise, and higher resolutions will lead to more resource consuming processing. The atlas shipped with Constellation is based on meshes of 32492 vertices per hemisphere, the ones used for the Human Connectome Project (HCP) preprocessings.
Thus in summary we have 3 different template meshes and resampling methods for it:
Diffusion data have to undergo a fiber tracking algorithm, which will produce either fiber tracts geometry reconstruction, or connectivity matrices. Constellation has been tested with two different tractography software: Connectomist2 and FSL probtrackx.
Other software, like MRtrix, Diffusion Toolkit / Trackvis or DiPy for instance, might also work, but have not been tested yet. Fiber tracts or connectivity matrices formats must be readable by Constellation, or converted to formats that can be read. The Trackvis format (.trk) is supported and is a convenient way to import tracts.
However all these software have not been validated yet for Constellation, and will probably require some additional work to handle not only file formats, but also coordinates systems and spatial transformations between fiber tracts and T1 images, and maybe adapt a few parameters.
Connectomist 2 should not be confused with the old Connectomist, the diffusion and tracking toolbox for BrainVISA, which is obsolete, deprecated, unmaintained, and has been removed from the BrainVISA distribution since BrainVISA version 4.6. Connectomist2, the successor, is also developed in Neurospin/France, by C. Poupon (a different team), but has not been released publicly so far.
Connectomist2 has to be run for each subject data. It can perform streamline tractography, using either deterministric or probabilistric tracking, using a given number of seeds in each brain voxel. Tracking is generally performed from each voxel of the brain. Constellation has been mostly used using probabilistric tractography with 27 seeds per voxel. This process typically generates about 2Gbytes of tracts per subject.
The main outputs of Connecomist2 used by Constellation are:
FSL provides its diffusion MRI processing and tractography pipelines. The main parts are T1 MRI brain extraction, Bedpostx (local diffusion model fit), and probtrackx (fiber tracking). Probtrackx can process a given number of seeds per voxels (typically 5000), and can track either from the cortex (mode omatrix1) or from the whole brain (omatrix3). Expect significant computing time, especially for high resolution and large data, and mostly using omatrix3 mode (for instance processing the HCP data took us more than 2 months of computing for a single subject).
Optionally, to perform cleaner tracking, it can be a good idea to provide FSL with a brain mask where sulci have been removed, to prevent tracts from crossing sulci. This can be done by running BrainVISA / Morphologist, and removing the sulci skeleton of Morphologist from the brain mask. This is typically what Connectomist2 does.
Important: Constellation works on Freesurfer surfaces, so FSL connectivity has to be processed with Freesurfer surfaces as masks in probtrackx. This operating mode is documented in the FSL user guide.
The main outputs of FSL probtrackx used by Constellation are:
Constellation starts once these preprocessings have been achieved.
In this situation, we want to build a new group-based parcellation on a given group of subjects, not using predefined atlases.
There is some intra-subject processing, and an inter-subject (group) clustering. They correspond to two different pipelines, the intra-subject one should be used before the inter-subject part.
Depending on the dMRI tractography software used for input to Constellation, there are different ways to handle them. Consequently there are currently 2 intra-subject piplines in Constellation, one dealing with Connectomist tracts, the other with FSL connectivity matrices. They differ on the first steps, and then have common later steps.
Group clusterings and assiociated connectivity profiles can then be used to make a new group atlas for instance. Group or individual connectivity profiles obtained on the clusters can be used to build and study connectivity graphs...
This is the simplest and lightest way to work: you come with one or several subjects data, and get a parcellation for them, with labels compatible with the atlas.
There are also two steps: one is to run the intra-subject pipeline above to get individuel connectivity matrices, then use the following process:
Constellation clusters, or other surface-based regions actually, can be used to reduce connectivity information into a region-based connectivity matrix: