brainvisa.data modules¶
brainvisa.data.neuroData¶
This module contains classes used to define the signature of Brainvisa processes, that is to say a list of typed parameters.
The main class is Signature. It contains a list of parameters names and types.
Each parameter is an instance of a sublcass of Parameter:
Number: an integer or float number.Choice: a choice between several constant values.ListOfVector: a list of vectors of numbers.Matrix: a matrix of numbers.brainvisa.data.readdiskitem.ReadDiskItem: abrainvisa.data.neuroDiskItems.DiskItemas an input parameter.brainvisa.data.writediskitem.WriteDiskItem: abrainvisa.data.neuroDiskItems.DiskItemas an output parameter.
Example
>>> signature = Signature(
>>> 'input', ReadDiskItem( "Volume 4D", [ 'GIS Image', 'VIDA image' ] ),
>>> 'output', WriteDiskItem( 'Volume 4D', 'GIS Image' ),
>>> 'threshold', Number(),
>>> 'method', Choice( 'gt', 'ge', 'lt', 'le' )
>>> )
Matching graphical editors classes are defined in brainvisa.data.qt4gui.neuroDataGUI.
- Inheritance diagram:
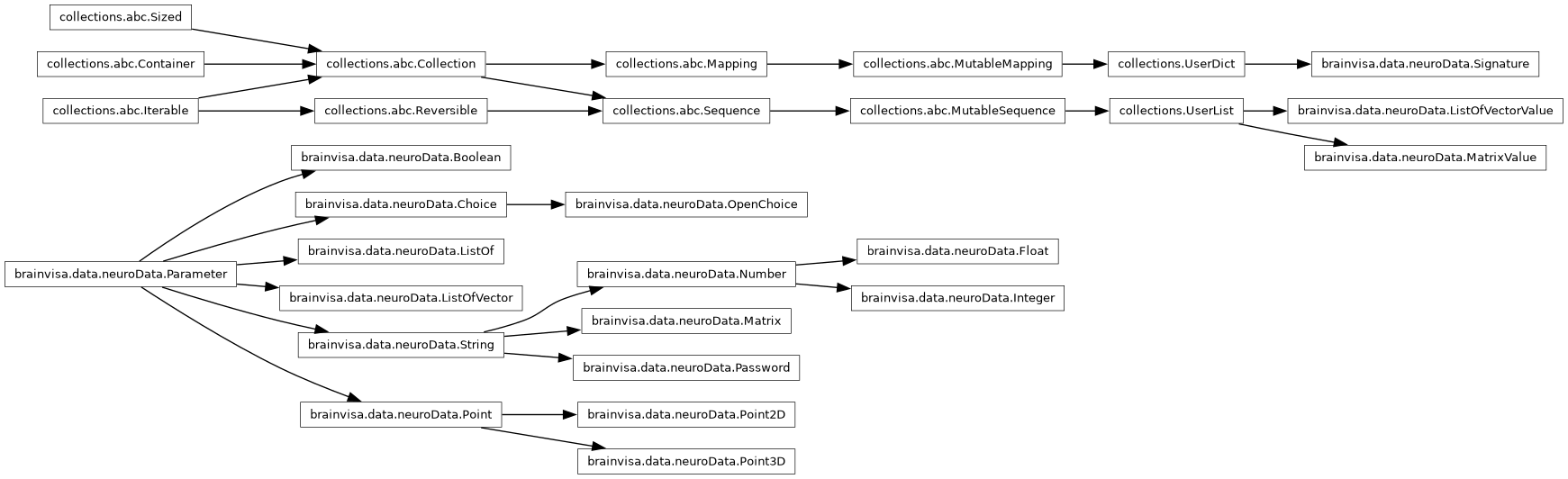
- Classes:
- class brainvisa.data.neuroData.Boolean(section=None, userLevel=0, mandatory=True, visible=True)[source]¶
Bases:
ParameterA choice between 2 values: True or False.
- class brainvisa.data.neuroData.Choice(*args, **kwargs)[source]¶
Bases:
ParameterA Choice parameter allows the user to choose a value among a set of possible values. This set is given as parameter to the constructor. Each value is associated to a label, which is the string shown in the graphical interface, possibly after a translation. That’s why a choice item can be a couple (label, value). When a choice item is a simple value, the label will be the string representation of the value (
label=unicode(value)).Examples
>>> c = Choice( 1, 2, 3 ) >>> c = Choice( ( 'first', 1 ), ( 'second', 2 ), ( 'third', 3 ) )
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- defaultValue()[source]¶
The default value for a choice parameter is the first of the list of choices or None if the list of choices is empty.
- findIndex(value)[source]¶
Finds the index of a value in the list of possible choices.
- Parameters:
value – could be the label or the associated value of the choice
- Return type:
integer
- Returns:
the index of the value if found, else -1.
- findValue(value)[source]¶
Finds a value in the list of choices that matches the one given in parameter. Raises
exceptions.KeyErrorif not found.
- setChoices(*args)[source]¶
Sets the list of possible values.
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- class brainvisa.data.neuroData.Float(*args, **kwargs)[source]¶
Bases:
NumberThis class represents a parameter that is float number.
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- class brainvisa.data.neuroData.Integer(*args, **kwargs)[source]¶
Bases:
NumberThis class represents a parameter that is an integer.
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- class brainvisa.data.neuroData.ListOf(contentType, allowNone=False, **kwargs)[source]¶
Bases:
ParameterThis parameter represents a list of elements of the same type.
- Attributes:
- Methods:
- Parameters:
contentType – type of the elements of the list.
- checkValue(name, value)[source]¶
Checks if the given value is valid for this parameter. The value should be a list or a tuple and each element should match the content type of the parameter. An exception is raised if these conditions are not met.
- findValue(value)[source]¶
Returns a suitable value for this parameter from the given value. If the value is None,
[]is returned. It it is a list, a new list with each value checked and possibly converted to the appropriate type is returned. If it is not a list (only one element), a list containing this element possibly converted to the appropriate type is returned.
- class brainvisa.data.neuroData.ListOfVector(length=None, **kwargs)[source]¶
Bases:
ParameterThis parameter expects a list of vectors value.
- Parameters:
length (int) – number of vectors.
- findValue(value)[source]¶
Returns a
ListOfVectorValuecreated from the given value, checking that the required length is respected.
- class brainvisa.data.neuroData.ListOfVectorValue(value, requiredLength=None)[source]¶
Bases:
UserListThis object represents a list of vectors.
- Attributes:
- data¶
The content of the list of vectors: a list of vectors, each vector is a list of number. The vectors can have different sizes.
- size¶
The number of vectors in the list.
- Parameters:
- class brainvisa.data.neuroData.Matrix(length=None, width=None, **kwargs)[source]¶
Bases:
StringThis parameter expects a matrix value.
- Parameters:
- findValue(value)[source]¶
Returns a
MatrixValuecreated from the given value, checking that the required dimensions are respected.
- class brainvisa.data.neuroData.MatrixValue(value, requiredLength=None, requiredWidth=None)[source]¶
Bases:
UserListThis object represents a matrix.
- Attributes:
- data(list)¶
Content of the matrix: a list of lines, each line is a list of number.
- size(tuple)¶
Dimension of the matrix (nb lines, nb columns)
- Parameters:
value (list) – content of the matrix: a list of lines, each line is a list of value. Each value should be a number of a string that can be converted to a number.
requiredLength (int) – required number of lines of the matrix. An exception is raised if this condition is not met.
requiredWidth (int) – required number of columns of the matrix. An exception is raised if this condition is not met.
- class brainvisa.data.neuroData.Number(*args, **kwargs)[source]¶
Bases:
StringThis class represents a parameter that is a number, integer of float.
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- class brainvisa.data.neuroData.OpenChoice(*args, **kwargs)[source]¶
Bases:
ChoiceAn OpenChoice enables to choose a value in the list of choice or a new value that is not in the list.
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- class brainvisa.data.neuroData.Parameter(section=None, userLevel=0, mandatory=True, visible=True)[source]¶
Bases:
objectThis class represents a type of parameter in a
Signature.- Attributes:
- mandatory¶
Boolean. Indicates if the parameter is mandatory, that is to say it must have a non null value. Default is True.
- userLevel¶
Integer. Indicates the minimum userLevel needed to see this parameter. Default is 0.
- databaseUserLevel¶
Integer. Indicates the minimum userLevel needed to allow database selection for this parameter (useful only for diskitems).
- browseUserLevel¶
Integer. Indicates the minimum userLevel needed to allow filesystem selection for this parameter (useful only for diskitems).
- linkParameterWithNonDefaultValue¶
Boolean. Indicates if the value of the parameter can be changed by the activation of a link between parameters even if the parameter has no more a default value (it has been changed by the user). Default is False.
- valueLinkedNotifier¶
soma.notification.Notifier. This notifier will notify its observers when a link to this parameter is activated.- Methods:
- checkReadable(value)[source]¶
This is a virtual function (returns always True) that could be overriden in derived class. It should return True if the value given in parameter is readable.
- checkValue(name, value)[source]¶
This functions check if the given value is valid for the parameter. If the value is not valid it raises an exception.
- editor(parent, name, context)[source]¶
Virtual function that can be overriden in derived class. The function should return an object that can be used to edit the value of the parameter. This one raises an exception saying that no editor exist for this type.
- getParameterized()[source]¶
Returns the
brainvisa.processes.Parameterizedobject associated to this parameter. Generally the Process that have this parameter in its signature.
- iterate(*args)[source]¶
Calls
findValue()on each value given in parameter. Returns the list of results.
- listEditor(parent, name, context)[source]¶
Virtual function that can be overriden in derived class. The function should return an object that can be used to edit a list of values for the parameter. This one raises an exception saying that no list editor exist for this type.
- setNameAndParameterized(name, parameterized)[source]¶
Stores a name and an associated
brainvisa.processes.Parameterizedobject in this parameter.
- class brainvisa.data.neuroData.Password(*args, **kwargs)[source]¶
Bases:
StringThis class represents a string parameter that is used as a Password.
- Parameters:
args (list) – list of possible value, each value can be a string or a tuple.
- class brainvisa.data.neuroData.Point(dimension=1, precision=None, **kwargs)[source]¶
Bases:
ParameterThis parameter type represents the coordinates of a point.
- Parameters:
- addLink(sourceParameterized, sourceParameter)[source]¶
Associates a specific link function between the source parameter and this parameter. When the source parameter changes, its value is stored in this object.
- Parameters:
sourceParameterized –
brainvisa.processes.Parameterizedobject that contains the parameters in its signaturesourceParameter –
Parameterobject that is the source of the link
- class brainvisa.data.neuroData.Point2D(dimension=2, precision=None, **kwargs)[source]¶
Bases:
PointPointin a two dimensions space.
- class brainvisa.data.neuroData.Point3D(dimension=3, precision=None, **kwargs)[source]¶
Bases:
PointPointin a three dimensions space.- Parameters:
- add3DLink(sourceParameterized, sourceParameter)[source]¶
Deprecated. Use
Point.addLink()instead.
- class brainvisa.data.neuroData.Signature(*params)[source]¶
Bases:
UserDictA list of parameters with the name and the type of each parameter.
This object can be used as a dictionary whose keys are the name of the parameters and the values are the types of the parameters. But unlike a python dictionary, the keys are ordered according their insertion in the signature object.
The constructor expects a list formatted as follow: parameter1_name (string), parameter1_type (
Parameter), …Each couple parameter_name, parameter_type defines a parameter. The parameter name is a string which must also be a Python variable name, so it can contain only letters (uppercase or lowercase), digits or underscore character. Moreover some Python reserved word are forbidden (and, assert, break, class, continue, def, del, elif, else, except, exec, finally, for, from, global, if, import, in, print, is, lambda, not ,or , pass, raise, return, try, why). The parameter type is an instance of
Parameterclass and indicates the supported value type for this parameter.- deepCopy()[source]¶
Returns a deep copy of the current signature. Both the parameters names and list of parameters types are duplicated.
- iteritems()[source]¶
iterates over a list of tuple (parameter_name, parameter_type) for each parameter.
brainvisa.data.neuroDiskItems¶
This module contains classes defining data items that are called diskItems in Brainvisa. These diskItems are associated to files that store data on a filesystem, and attributes that are used to index the data in a database.
The main class in this module is DiskItem. This class is derived into two sub-classes File for the diskitems that are stored as files and Directory for the diskitems that represent directories.
A diskItem has a type indicating what the data represents (a Volume, a T1 MRI, fMRI data…) and a format indicating the file format used to write the data in files.
The class DiskItemType represents data types.
The class Format represents file formats.
Two general formats are defined in this module as global variables:
- brainvisa.data.neuroDiskItems.directoryFormat¶
This format matches any directory.
- brainvisa.data.neuroDiskItems.fileFormat¶
This format matches any file.
The available types and formats in Brainvisa ontology are defined in each toolbox in python files under the types directory.
They are loaded at Brainvisa startup using the function readTypes() and stored in global maps:
- brainvisa.data.neuroDiskItems.formats¶
A global map which associates each format id to the matching object
Format.
- brainvisa.data.neuroDiskItems.formatLists¶
A global map which associates each list of formats id to the matching object
NamedFormatList.
- brainvisa.data.neuroDiskItems.diskItemTypes¶
A global map which associates each type id to the matching object
DiskItemType
The following function are available to get a format or a type object from its id:
This module also defines classes for temporary files and directories: TemporaryDiskItem, TemporaryDirectory.
The function getTemporary() enables to create a new temporary item.
All temporary files and directories are written in Brainvisa global temporary directory which can be choosen in Brainvisa options. The diskitem corresponding to this global directory is stored in a global variable:
- brainvisa.data.neuroDiskItems.globalTmpDir¶
It is the default parent diskItem for temporary diskitems.
- Inheritance diagram:
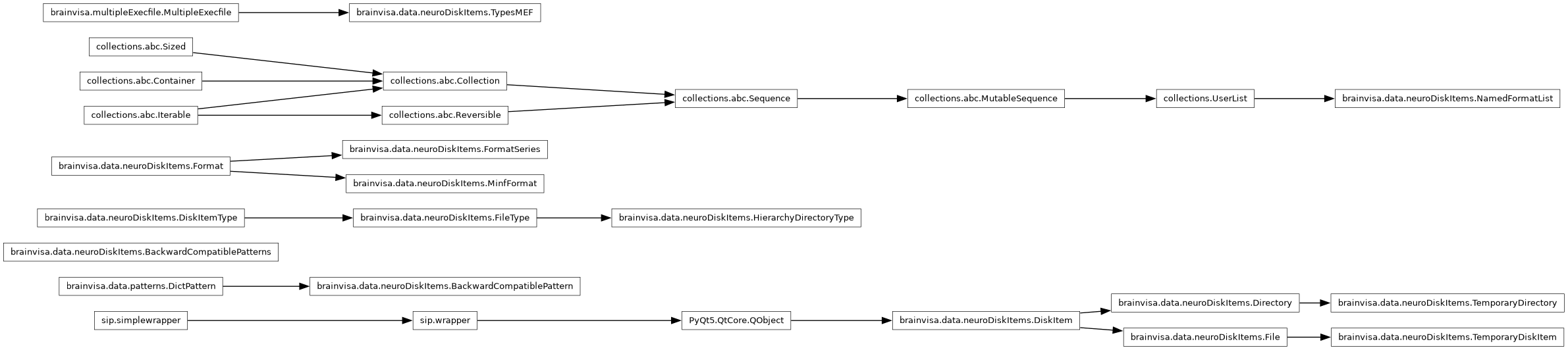
- Classes:
- class brainvisa.data.neuroDiskItems.BackwardCompatiblePattern(pattern)[source]¶
Bases:
DictPatternThis class represents a file pattern.
The pattern is described with an expression like
f|*.jpg. The first character (before the|) indicates if the pattern recognizes files (f) or directories (d). This part is optional. The rest of the pattern is a simple regular expression that describes the file name. It can contain*character to replace any character and#character to replace numbers. See the parent classbrainvisa.data.patterns.DictPatternfor more details about the patterns.A method
match()enables to check if aDiskItemfile name matches this format pattern.- match(diskItem)[source]¶
Checks if the diskitem file name matches the pattern.
- Parameters:
diskItem – The
DiskItemwhose file format is checked.- Returns:
a dictionary containing the value found in the diskItem file name for each named expression of the pattern. The
*character in the pattern is associated to afilename_variablekey in the result dictionary. The#character in the pattern is associated to aname_seriekey in the result dictionary.Noneis returned if the diskitem file name doesn’t match the pattern.
- unmatch(diskItem, matchResult, force=False)[source]¶
The opposite of
match()method: the matching string is found from a match result and a dictionary of attributes values.- Parameters:
matchResult – dictionary which associates a value to each named expression of the pattern.
dict – dictionary which associates a value to each attribute name of the pattern.
force (bool) – If True default values are set in the match result for
filename_variableandname_serieattributes.
- Return type:
string
- Returns:
The rebuilt matching string.
- class brainvisa.data.neuroDiskItems.BackwardCompatiblePatterns(patterns)[source]¶
Bases:
objectThis class represents several file patterns.
Each pattern is a
BackwardCompatiblePattern.- Parameters:
patterns – a string, a list of string, or a list of
BackwardCompatiblePattern. They are used to create the internal list ofBackwardCompatiblePattern.
- fileOrDirectory()[source]¶
Checks if all the patterns match files or directories. If they all match the same type (file or directory), returns this type.
- match(diskItem, returnPosition=0)[source]¶
Checks if the diskitem matches one of the patterns of this
BackwardCompatiblePatterns.- Parameters:
diskItem – The diskitem which files names should match the patterns.
returnPosition (bool) – if True, the index of the matching pattern is returned as well as the match result.
- Returns:
the match result or a tuple (match result, index) if
returnPositionis True.
- class brainvisa.data.neuroDiskItems.Directory(name, parent)[source]¶
Bases:
DiskItemThis class represents a directory, that is to say a diskItem that can contain other diskItems.
- class brainvisa.data.neuroDiskItems.DiskItem(name, parent)[source]¶
Bases:
QObjectThis class represents data stored in one or several files on a filesystem. It can have additional information stored in attributes and may be indexed in a Brainvisa database.
A diskItem can have hierarchy attributes (comes from Brainvisa ontology), minf attributes (that are stored in a .minf file), and possibly other attributes. Several methods enable to access the values of the attributes:
getHierarchy(),getNonHierarchy(),attributes(),globalAttributes(),hierarchyAttributes(),localAttributes(),minf(). The attributes of a diskItem can also be requested using the dictionary notationd["attribute_name"], which callsget()method.A diskItem can be identified by a unique identifier called uuid. This uuid is stored in minf attributes and it is an instance of
soma.uuid.Uuidclass.Several methods enable to request the name of the files associated to the diskItem:
fullPath(),fullName(),fullPaths(),fullPathSerie(),fullPathsSerie().- Attributes:
- name¶
name of the diskItem, generally the filename of the first file.
- parent¶
a parent diskItem, generally the diskItem associated to the directory that contains the data files of this diskItem.
- type¶
Data type of the diskItem, indicating the meaning of the data. It is an instance of
DiskItemType.
- Methods:
- attributes()[source]¶
Returns a dictionary containing the name and value of all the attributes associated to this diskItem.
- clearMinf(saveMinf=True)[source]¶
Deletes all minf attributes. Also removes the minf file if saveMinf is True.
- clone()[source]¶
Returns a deep copy of the current diskItem object. The associated files are not copied.
- copyAttributes(other)[source]¶
Copy all the attributes of the given diskItem in the current diskItem.
- Parameters:
other – the
DiskItemwhose attributes will be copied.
- createParentDirectory()[source]¶
According to the file path of the diskItem, creates the directory that should contain the files if it doesn’t exist.
- distance(other)[source]¶
Returns a value that represents a sort of distance between two DiskItems. The distance is not a number but distances can be sorted, it is a tuple of numbers.
- existingFiles()[source]¶
Returns all files associated to this diskitem, that really exist plus its minf file if it exists.
- fileName(index=0, withQueryString=True)[source]¶
Returns the filename of the file number index in the list of files associated to this diskItem. The absolute files paths are generally stored in diskItems. So this function is equivalent to
fullPath().
- fileNameSerie(serie, index=0)[source]¶
This function can be used only on diskItems that are a serie of data. For example a serie of 3D volumes, each volume being stored in two files .ima and .dim (GIS format). Returns the name of the index number file of the serie number item of the serie. The absolute files paths are generally stored in diskItems. So this function is equivalent to
fullPathSerie().
- fileNames(withQueryString=True)[source]¶
Returns the list of filenames of the files associated to this diskItem. The absolute files paths are generally stored in diskItems. So this function is equivalent to
fullPaths().
- fileNamesSerie(serie)[source]¶
Returns all the files of one item of the serie. The absolute files paths are generally stored in diskItems. So this function is equivalent to
fullPathsSerie().- Parameters:
serie (int) – index of the item in the serie.
- findFormat(amongFormats=None)[source]¶
Find the format of this diskItem : the format whose pattern matches this diskitem’s filename. Does nothing if this item has already a format. Doesn’t take into account format whose pattern matches any filename (*). Stops when a matching format is found :
item name is modified (prefix and suffix linked to the format are deleted)
item list of files is modified accoding to format patterns
the format is applied to the item
setFormatAndTypeAttributes()method is applied.
- fullPath(index=0, withQueryString=True)[source]¶
Returns the absolute file name of the index number file of the diskItem.
- fullPathSerie(serie, index=0)[source]¶
This function can be used only on a diskItem that is a serie of data. For example a serie of 3D volumes, each volume being stored in two files .ima and .dim (GIS format). Returns the absolute file name of the index number file of the serie number item of the serie.
- fullPaths(withQueryString=True)[source]¶
Returns the absolute file names of the all the files of the diskItem.
- fullPathsSerie(serie)[source]¶
This function can be used only on a diskItem that is a serie of data. For example a serie of 3D volumes, each volume being stored in two files .ima and .dim (GIS format). Returns all the absolute file names of the serie number item of the serie.
- Parameters:
serie (int) – index of the item in the serie.
- get(attrName, default=None, search_header=False)[source]¶
Gets the value of an attribute. If the attribute is not found in the attributes stored in this diskItem object, it can be searched for in data file header using
aimsFileInfo()function if the option search_header is True.Warning
If search_header is True, the method can take more time to execute than
getHierarchy()andgetNonHierarchy()when the attribute is not found because it reads the header of the file on the filesystem.- Parameters:
attrName (string) – name of the attribute
default – value returned if the attribute is not set in this diskItem.
- getHierarchy(attrName, default=None)[source]¶
Gets the attribute from the global attributes or the local attributes or the parent hierarchy attributes.
- Parameters:
attrName (string) – name of the attribute
default – value returned if the attribute is not found.
- getInTree(attrPath, default=None, separator='.')[source]¶
This function could be used to get an attribute value from an object that is an attribute value of a diskItem.
d.getInTree("attr1.attr2...") <=> d.get(attr1).get(attr2)...
- Parameters:
attrPath (string) – the attributes path, each attribute is separated by a separator character
default – default value is the attribute value is not found
separator (string) – character separator used to separate the different attributes in the attributes path.
- getNonHierarchy(attrName, default=None)[source]¶
Gets the attribute value from the attributes written in minf file or in other attributes or in parent non hierarchy attributes.
- Parameters:
attrName (string) – name of the attribute
default – value returned if the attribute is not found.
- getResolutionDimensions()[source]¶
Get the multi resolution dimensions for a . If the file is not an image, None is returned. :return: The current resolution level of
DiskItem.
- globalAttributes()[source]¶
Returns a dictionary containing the name and value of the global attributes. They may come from parent diskItem and are transmitted to child diskItem. The global attributes are the attributes coming from data ontoloy.
- isReadable()[source]¶
Returns True if all the files associated to this diskItem exist and are readable.
- isTemporary()[source]¶
Returns True if it is a temporary diskItem, that is to say its files will be automatically deleted when its is no more referenced.
- isWriteable()[source]¶
Returns True if all the files associated to this diskItem which exist are readable and writable and if they do not exist if the parent directories exist and are writable and executable.
- localAttributes()[source]¶
Returns a dictionary containing the name and value of the local attributes. The local attributes are valid only for this diskItem, they are not transmitted to children diskItems. For example the attribute
name_seriethat indicates the list of numbers of a series of files that store the data, is a local attribute.
- minfFileName()[source]¶
Returns the name of the minf file associated to this diskItem. It is generally the name of the main file of the diskItem + the .minf extension.
- modificationHash()[source]¶
Return a value that can be used to assess modification of this DiskItem. Two calls to modificationHash will return the same value if and only if all files in the DiskItem have not changed. Note that the contents of the files are not read, the modification hash rely only on os.stat.
This method uses the funciton
modificationHashOrEmpty()to create a hash code for each file.
- priority()[source]¶
Returns the value of priority attribute if found else the parent diskItem priority, else the default priority attribute, else 0.
This priority is an attribute associated to the ontology rule that enabled to identify the diskItem.
- readAndUpdateMinf()[source]¶
Reads the content of the minf file and updates the minf attribute dictionary accordingly.
- relativePath(index=0)[source]¶
Gets the file path of this diskitem, relatively to the path its database directory. If there is no database information in the attributes it returns the full path of the diskItem.
- removeMinf(attrName, saveMinf=True)[source]¶
Remove the attribute from minf attributes.
- Parameters:
attrName (string) – name of the attribute to remove
saveMinf (bool) – if True, the minf attributes are saved in the minf file.
- resolutionLevel()[source]¶
Get the current resolution level of a DiskItem. The resolution level is read from the querystring. :return: The current resolution level of DiskItem.
- saveMinf(overrideMinfContent=None)[source]¶
Writes minf attributes or the content of overrideMifContent if given to the minf file.
- setFormatAndTypeAttributes(writeOnly=0)[source]¶
Adds the diskItem format attributes to its other attributes, and the diskItem types attributes to its minf attributes.
- Parameters:
writeOnly (bool) – if False the minf file is read to update the dictionary of minf attributes.
- Returns:
current disktem
- setMinf(attrName, value, saveMinf=True)[source]¶
Adds this attribute to the minf attributes of the diskItem.
- Parameters:
attrName (string) – name of the attribute
value – value sets for the attribute
saveMinf (bool) – if True the modified attributes are written to the diskItem minf file.
- setPriority(newPriority, priorityOffset=0)[source]¶
Sets a value for the priority attribute:
newPriority + priorityOffset.
- setResolutionLevel(resolution_level)[source]¶
Sets the resolution level in the queryString of a DiskItem. It allows to use a specific resolution for multi resolution data. Once set, the fullPath of the DiskItem becomes ‘/dir/name.ext?resolution_level=<resolution_level>’
- Parameters:
resolution_level – The DiskItem resolution level
to use.
- updateMinf(dict, saveMinf=True)[source]¶
Adds new attributes to the minf attributes of the diskItem and possibly save the attributes in a minf file.
Note: it is not possible to remove attributes using this function, as it only changes the values of the attributes given in the dict.
- Parameters:
dict – dictionary containing the attributes that will be added to this diskItem minf attributes.
saveMinf (bool) – if True the minf attributes will be saved in the minf file of the diskItem.
- class brainvisa.data.neuroDiskItems.DiskItemType(typeName, *args, **kwargs)[source]¶
Bases:
objectThis class represents a data type. It is used to define the type attribute of
DiskItemobjects.The types are defined hierarchically, that’s why a type can have a parent type and a method
DiskItemType.isA()enables to request if a type is derived from another type.- Attributes:
- name¶
Name of the Type. For example T1 MRI.
- fileName¶
Name of the python file that contains the definition of this type.
- id¶
Identifier associated to this type.
- parent¶
As types are defined hierarchically, a type can have a parent type.
- toolbox¶
Name of the toolbox that defined the type, default is ‘axon’ (no toolbox).
- Methods:
- Parameters:
typeName (string) – name of the type
parent – can be the name of the parent. If it already registered, it will be found from its name.
attributes – dictionary containing attributes associated to this type.
- inheritanceLevels(diskItemType)[source]¶
Returns -1 if the given diskItem type is not a parent (direct or indirect) of the current diskItem, else the number of levels between the given diskItem type and the current diskItem type.
- isA(diskItemType)[source]¶
Returns True if the given diskItem type is a parent (direct or indirect) of the current diskItem.
- levels()[source]¶
Returns hierarchy levels from the current diskItem. Each level is a tuple that contains an integer and a diskItem type.
- parents()[source]¶
Returns the list of parents of this diskItem, that is to say its parent and its parent’s parents.
- class brainvisa.data.neuroDiskItems.File(name, parent)[source]¶
Bases:
DiskItemThis class represents a diskItem that cannot contain other diskItems (it is not a directory).
- class brainvisa.data.neuroDiskItems.FileType(typeName, *args, **kwargs)[source]¶
Bases:
DiskItemTypeThis class represents a type for a DiskItem associated to files (a
File). It is used to define new data types in brainvisa ontology files.It can store file formats associated to this type.
- Parameters:
typeName (string) – name of the type
formats – list of file formats associated to this type of data.
minfAttributes – dictionary containing attributes associated to this type.
- sameContent(other, as_dict=False)[source]¶
Returns True if the two file types are instances of the same class and have the same list of formats and the same parent.
The formats and parents are compared using the function
sameContent().
- class brainvisa.data.neuroDiskItems.Format(*args, **kwargs)[source]¶
Bases:
objectThis class represents a data file format. It is used to define the format attribute of
DiskItemobjects. It is also used to define new formats in brainvisa ontology files.- Attributes:
- name¶
Name of the format. For example GIS image.
- fileName¶
Name of the python file that contains the definition of this format.
- id¶
Identifier associated to this format.
- patterns¶
BackwardCompatiblePatternsdescribing the files patterns associated to this format. Example:f|*.ima, f|*.dim.
- toolbox¶
Name of the toolbox that defined the format, default is ‘axon’ (no toolbox).
- Methods:
- Parameters:
formatName (string) – name of the format.
patterns – string or
BackwardCompatiblePatternsdescribing the files patterns associated to this format.attributes – dictionary of attributes associated to this format. For example, an attribute compressed = type_of_compression is associated to the compressed files formats.
exclusive (bool) – if True, a file should match only this pattern to match the format. Hardly ever used.
ignoreExclusive (bool) – if True, this format will be ignored when exclusivity of a format is checked. Hardly ever used.
- fileOrDirectory()[source]¶
Returns the class of diskItem that this format matches :
FileorDirectory.
- formatedName(item, matchResult)[source]¶
Returns the name of the diskItem that matches the format without format extension.
- getPatterns()[source]¶
Returns a
BackwardCompatiblePatternsrepresenting the files patterns for this format.
- match(item, returnPosition=0, ignoreExclusive=0)[source]¶
Checks if the diskItem files match the format patterns.
- Parameters:
- Returns:
a dictionary containing the matched variables of the patterns or a tuple containing this dictionary and the position of the match if returnPosition was True.
- setAttributes(item, writeOnly=0)[source]¶
Adds the attributes of this format to the diskItem other attributes
- class brainvisa.data.neuroDiskItems.FormatSeries(*args, **kwargs)[source]¶
Bases:
FormatThis class represents the format of a serie of diskItems with the same format. For example, a serie of images in Analyse format. Such an object can be created using the function
changeToFormatSeries().- Attributes:
- baseFormat¶
The format of each element of the serie.
- Methods:
- Parameters:
formatName (string) – name of the format.
patterns – string or
BackwardCompatiblePatternsdescribing the files patterns associated to this format.attributes – dictionary of attributes associated to this format. For example, an attribute compressed = type_of_compression is associated to the compressed files formats.
exclusive (bool) – if True, a file should match only this pattern to match the format. Hardly ever used.
ignoreExclusive (bool) – if True, this format will be ignored when exclusivity of a format is checked. Hardly ever used.
- fileOrDirectory()[source]¶
Calls the
Format.fileOrDirectory()method of the base format.
- formatedName(item, matchResult)[source]¶
Calls the
Format.formatedName()method of the base format.
- getPatterns()[source]¶
Calls the
Format.getPatterns()method of the base format.
- group(groupedItem, matchedItem, position=0, matchRule=None)[source]¶
Groups files which are associated to the same diskitem.
- match(*args, **kwargs)[source]¶
Calls the
Format.match()method of the base format.
- postProcessing(item)[source]¶
Sorts and removes doubloons in the list of numbers in the
name_serieattribute of the item.
- unmatch(*args, **kwargs)[source]¶
Calls the
Format.unmatch()method of the base format.
- class brainvisa.data.neuroDiskItems.HierarchyDirectoryType(typeName, *args, **kwargs)[source]¶
Bases:
FileTypeThis type represents a directory which represents data in Brainvisa ontology. It is associated to the format
directoryFormat.- Parameters:
typeName (string) – name of the type
formats – list of file formats associated to this type of data.
minfAttributes – dictionary containing attributes associated to this type.
- class brainvisa.data.neuroDiskItems.MinfFormat(*args, **kwargs)[source]¶
Bases:
FormatBase class for the file formats that uses the minf format.
- Parameters:
formatName (string) – name of the format.
patterns – string or
BackwardCompatiblePatternsdescribing the files patterns associated to this format.attributes – dictionary of attributes associated to this format. For example, an attribute compressed = type_of_compression is associated to the compressed files formats.
exclusive (bool) – if True, a file should match only this pattern to match the format. Hardly ever used.
ignoreExclusive (bool) – if True, this format will be ignored when exclusivity of a format is checked. Hardly ever used.
- class brainvisa.data.neuroDiskItems.NamedFormatList(name, data)[source]¶
Bases:
UserListThis class represents a list of formats which have a name. This object can be used as a list.
- name¶
Name of the list of formats.
- data¶
List of formats.
- fileName¶
Name of the file where this list of formats is defined.
- class brainvisa.data.neuroDiskItems.TemporaryDirectory(name, parent)[source]¶
Bases:
DirectoryThis class represents a temporary directory.
It will be deleted when Brainvisa closes.
The deletion of temporary files and directories is managed by a
brainvisa.data.temporary.TemporaryFileManager.
- class brainvisa.data.neuroDiskItems.TemporaryDiskItem(name, parent)[source]¶
Bases:
FileThis class represents a temporary diskitem.
The associated files are automatically deleted when the corresponding object is no more used and so garbage collected.
The deletion of temporary files and directories is managed by a
brainvisa.data.temporary.TemporaryFileManager.
- class brainvisa.data.neuroDiskItems.TypesMEF[source]¶
Bases:
MultipleExecfileThis class enables to read Brainvisa ontology files which contain the definition of formats and types.
A global instance of this object
mefis used to load types and formats at Brainvisa startup.The Brainvisa types files will define new formats with
Format, new types withFileTypeand new format lists withcreateFormatList().With the
brainvisa.multipleExecfile.MultipleExecfile.localDict, callback methods are associated to the functions that will be called in the types files:create_format(),create_format_list(),create_type().- create_format(*args, **kwargs)[source]¶
This method is called when a new format is created in one of the executed files. The toolbox, module and fileName attributes of the new
Formatare set.- Parameters:
args – The arguments will be passed to the constructor of
Format.- Returns:
The new format.
- create_format_list(*args, **kwargs)[source]¶
This method is called when a new formats list is created in one of the executed files. The toolbox, module and fileName attributes of the new
NamedFormatListare set.- Parameters:
args – The arguments will be passed to the function
createFormatList().- Returns:
The new formats list.
- create_format_serie(*args, **kwargs)[source]¶
This method is called when a new formats list is created in one of the executed files. The toolbox, module and fileName attributes of the new
FormatSeriesare set.- Parameters:
args – The arguments will be passed to the function
changeToFormatSeries().- Returns:
The new format series.
- create_hie_dir_type(*args, **kwargs)[source]¶
This method is called when a new type is created in one of the executed files. The toolbox, module and fileName attributes of the new
HierarchyDirectoryTypeare set.- Parameters:
args – The arguments will be passed to the constructor of
HierarchyDirectoryType.- Returns:
The new type.
- brainvisa.data.neuroDiskItems.aimsFileInfo(fileName)[source]¶
Reads the header of the file fileName and returns its attributes as a dictionary.
- brainvisa.data.neuroDiskItems.changeToFormatSeries(format)[source]¶
Gets the suited format for a serie of item in the given format.
- brainvisa.data.neuroDiskItems.createFormatList(listName, formats=[])[source]¶
Creates a new
NamedFormatListand stores it in a global list of formats lists.- Parameters:
listName (string) – the name of the list
formats (list) – the list of formats ids (string)
- Returns:
the new
NamedFormatListcontaining a list ofFormatobjects.
- brainvisa.data.neuroDiskItems.expand_name_serie(text, number)[source]¶
Replaces
#character in text by number.
- brainvisa.data.neuroDiskItems.getAllDiskItemTypes()[source]¶
Gets all registered
DiskItemType.
- brainvisa.data.neuroDiskItems.getDiskItemType(item)[source]¶
Gets the
DiskItemTypewhose id is the given item.
- brainvisa.data.neuroDiskItems.getFormat(item, default=<undefined>)[source]¶
Gets the format whose id is the given item.
- brainvisa.data.neuroDiskItems.getFormats(formats)[source]¶
Returns a list of
Formator aNamedFormatListcorresponding to the given parameter. The parameter can be a string or a list of string matching format names.
- brainvisa.data.neuroDiskItems.getResolutionsFromItems(items)[source]¶
Get the multi resolutions string for common resolution levels of a list of items.
- Parameters:
items – list of py:class:DiskItem.
- Returns:
The list of common resolutions.
- brainvisa.data.neuroDiskItems.getTemporary(format, diskItemType=None, parent=None, name=None, suffix=None, prefix=None)[source]¶
Creates a new temporary diskitem. It will be automatically deleted when there is no more references on it or when Brainvisa closes.
- Parameters:
format – format of the diskitem. If the format correspond to a directory, a
TemporaryDirectorywill be created, else aTemporaryDiskItemwill be created.diskItemType – type of the diskItem.
parent – parent diskitem: directory which contains the diskItem. By default it will be Brainvisa global temporary directory whose path can be choosen in the options. The global temporary directory diskitem is defined in the global variable
globalTmpDir.name – filename for the diskitem. By default, a new name is generated by the
brainvisa.data.temporary.TemporaryFileManager.
- Returns:
a
TemporaryDirectoryor aTemporaryDiskItem.
- brainvisa.data.neuroDiskItems.isSameDiskItemType(base, ref)[source]¶
Returns True if ref is a parent of base.
- brainvisa.data.neuroDiskItems.modificationHashOrEmpty(f)[source]¶
Returns a tuple containing information about the file from
os.lstat(). Returns an empty tuple if an exception occurs.
- brainvisa.data.neuroDiskItems.readTypes()[source]¶
This function loads types and formats by reading Brainvisa ontology types files.
- brainvisa.data.neuroDiskItems.registerFormat(format)[source]¶
Stores the format in a global list of formats.
- brainvisa.data.neuroDiskItems.reloadTypes()[source]¶
This function reinitializes the global variables used to store the types and formats before calling
readTypes().
- brainvisa.data.neuroDiskItems.sameContent(a, b, as_dict=False)[source]¶
Checks if a and b have the same content.
if as_dict it True, b may be a dict when a is an instance: b is an instance.__dict__ for example.
If the two objects are lists, the function is called on each element of the list. If the first object has a method named sameContent, it is called. Else, the result of a comparison with == operator is returned.
brainvisa.data.neuroHierarchy¶
This module defines functions to load Brainvisa databases.
The databases objects are stored in a global variable:
- brainvisa.data.neuroHierarchy.databases¶
brainvisa.data.sqlFSODatabase.SQLDatabasesobject which contains the loaded Brainvisa databases.
This object is created with the function initializeDatabases().
Then each each database can be loaded with the function openDatabases().
An object brainvisa.data.sqlFSODatabase.SQLDatabase is created for each database selected in Brainvisa options.
The function hierarchies() enables to get the list of databases objects.
- brainvisa.data.neuroHierarchy.hierarchies()[source]¶
Returns a list of
brainvisa.data.sqlFSODatabase.SQLDatabaseobjects representing the databases currently loaded in Brainvisa.
- brainvisa.data.neuroHierarchy.initializeDatabases()[source]¶
Creates the object
databaseswhich is an instance ofbrainvisa.data.sqlFSODatabase.SQLDatabases. It will contain all loaded databases.
- brainvisa.data.neuroHierarchy.openDatabases()[source]¶
Loads databases which are selected in Brainvisa options. For each database, an object
brainvisa.data.sqlFSODatabase.SQLDatabaseis created. The new database objects are added todatabases, any existing database in this object is previously removed.Warning messages may be displayed if a database is readonly or uses a deprecated ontology (brainvisa-3.0).
brainvisa.data.fileSystemOntology¶
This module contains classes defining Brainvisa ontology rules to manage a database directory.
In Brainvisa ontology, adding to the types and formats definition, it is possible to define rules associating a data type and a path in a location and filename in a database directory. The classes in this module enable to define such rules.
These rules describe the organization of data in the database filesystem. Thanks to this description, the name and place of a file allows to guess its type and some information about it, as for example the center, subject or acquisition associated to this data. It also makes it possible to write data in the database using the same rules, so the information can be retrieved when the data is reloaded later.
These ontology files that we called hierarchy files in Brainvisa are located in brainvisa/hierarchies directory and in the hierarchies directory of each toolbox. BrainVISA can use several hierarchies whose files are grouped in a directory named as the hierarchy.
The main class in this module is FileSystemOntology.
It represents a Brainvisa hierarchy, a set of rules that associate data types and data organization on the filesystem.
A rule is represented by the class ScannerRule.
Several classes inheriting from ScannerRuleBuilder are used to associate ontology attributes to a rule.
- Inheritance diagram:

- Classes:
- class brainvisa.data.fileSystemOntology.DeclareAttributes(*attributes)[source]¶
Bases:
ScannerRuleBuilderLists attributes which should be added to the database columns, with no default value. :param string attributes: name of the attributes
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.DirectoryScanner(scannerRules)[source]¶
Bases:
objectThis object contains a list of
ScannerRuledescribing the content of a directory.- Attributes:
- rules¶
The list of
ScannerRuledescribing the content of the directory.
- possibleTypes¶
Dictionary containing the data types associated to its rules as keys. Values are always 1.
- Methods:
- class brainvisa.data.fileSystemOntology.FileSystemOntology(source, directories)[source]¶
Bases:
objectThis class represents a Brainvisa hierarchy, that is to say a set of rules associating data types and filenames.
The right way to use this class is to use the
get()method to get an instance of this class for a specific hierarchy in order to create only one instance for each hierarchy.- Attributes:
- name¶
The name of the hierarchy. It is the name of the directory containing the hierachy files, under the hierarchies directory.
- source¶
List of source paths for this hierarchy. Indeed, the hierarchy files can be in several directories: in the main Brainvisa directory and in each toolbox directory.
- content¶
Content of the hierarchy as it is described in the hierarchy files.
- typeToPatterns¶
Map associating each data type (
brainvisa.data.neuroDiskItems.FileType) with a list of rules (ScannerRule).
- lastModification¶
Date of last modification of the hierarchy files. This enables to detect ontology changes and to offer the user to update his databases.
- Methods:
- static get(source)[source]¶
Satic factory for creation of FileSystemOntology instances. The source can be:
The name of one of the FSO directories located in one of the “hierarchies” directories of neuroConfig.fileSystemOntologiesPath (for example ‘brainvisa-3.2.0’ is the main FSO).
The name of any FSO directory.
The name of an old-style (prior to version 3.1) hierarchy file.
- static getOntologiesNames()[source]¶
Lists all the ontologies names found in fileSystemOntologiesPath.
- printFormats(file=<_io.TextIOWrapper name='<stdout>' mode='w' encoding='utf-8'>)[source]¶
Prints information about formats.
- class brainvisa.data.fileSystemOntology.ScannerRule(pattern)[source]¶
Bases:
objectThis class represents a hierarchy rule. It associates a filename pattern and ontology attributes.
- Attributes:
- pattern¶
The filename pattern for this rule. It is an instance of
brainvisa.data.neuroDiskItems.BackwardCompatiblePattern.
- globalAttributes¶
List of global attributes names and values (tuples). Global attributes are added with a
SetAttrbuilder.
- localAttributes¶
List of local attributes names and values (tuples). Local attributes are added with a
SetWeakAttrbuilder.
- defaultAttributesValues¶
Dictionary associating attribute names and default values. It can be added with a
SetDefaultAttributeValuebuilder.
- type¶
Data type associated to this rule. Instance of
brainvisa.data.neuroDiskItems.DiskItemType. It can be added with aSetTypebuilder.
- formats¶
List of file formats associated to this rule, each format is an instance of
brainvisa.data.neuroDiskItems.Format. It can be added with aSetFormatsbuilder.
- scanner¶
When the rule pattern matches a directory, it can contain other elements. In this case, this attribute is a
DirectoryScannerand it contains other rules describing the content of the directory.
- priority¶
This attribute can be added with a
SetPrioritybuilder.
- priorityOffset¶
This attribute can be added with a
SetPriorityOffsetbuilder.
- fileNameAttribute¶
This attribute can be added with a
SetFileNameAttributeor aSetFileNameStrongAttributebuilder.
- fileNameAttributeIsWeak¶
This attribute can be added with a
SetFileNameAttributeor aSetFileNameStrongAttributebuilder.
- fileNameAttributeDefault¶
Default value for the filename attribute.
- nonMandatoryKeyAttributes¶
This attribute can be added with a
SetNonMandatoryKeyAttributebuilder.
- class brainvisa.data.fileSystemOntology.ScannerRuleBuilder[source]¶
Bases:
objectBase class for rule builders.
It defines a virtual method
build(). All derived class override this method.- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetAttr(*params)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.globalAttributesattribute of the current rule.- Parameters:
params – list of attribute name followed by its value.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetContent(*params)[source]¶
Bases:
ScannerRuleBuilderThis builder assumes that the current rule pattern is matches a directory. As a directory can contain other files, a new
DirectoryScanneris created and set as theScannerRule.scannerattribute of the current rule. This directory scanner contains the rules defined to describe the content of this directory.- Parameters:
params – list of rules describing the content of a directory: several filename patterns followed by associated rule builders.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetDefaultAttributeValue(attribute, value)[source]¶
Bases:
ScannerRuleBuilderThis builder adds a new attribute value in the
ScannerRule.defaultAttributesValuesmap of the current rule.- Parameters:
attribute (string) – name of the attribute
value (string) – default value of the attribute.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetFileNameAttribute(attribute, defaultValue=None)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.fileNameAttributeandScannerRule.fileNameAttributeDefaultattributes of the current rule. TheScannerRule.fileNameAttributeIsWeakattribute is set to 1.- Parameters:
attribute (string) – name of the attribute
defaultValue (string) – a default value for this attribute.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetFileNameStrongAttribute(attribute, defaultValue=None)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.fileNameAttributeandScannerRule.fileNameAttributeDefaultattributes of the current rule. TheScannerRule.fileNameAttributeIsWeakattribute is set to 0.- Parameters:
attribute (string) – name of the attribute
defaultValue (string) – a default value for this attribute.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetFormats(formats)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.formatsattribute of the current rule.- Parameters:
formats – list of formats names that will be associated to the current pattern.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetName(name)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.itemNameattribute of the current rule.- Parameters:
name (string) – a name that will be associated to any diskitem that match the rule.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetNonMandatoryKeyAttribute(*attributes)[source]¶
Bases:
ScannerRuleBuilderThis builder adds new attributes names in the
ScannerRule.nonMandatoryKeyAttributeslist of the current rule.- Parameters:
attributes – list of attributes names that are not mandatory key attributes.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetPriority(priority)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.priorityattribute of the current rule.- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetPriorityOffset(priorityOffset)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.priorityOffsetattribute of the current rule.- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- class brainvisa.data.fileSystemOntology.SetType(diskItemType)[source]¶
Bases:
ScannerRuleBuilderThis builder set the
ScannerRule.typeattribute of the current rule.- Parameters:
diskItemType (string) – data type.
- build(scannerRule)[source]¶
Sets its data type to the given scannerRule.
If the type has associated formats, the
ScannerRule.formatsis also updated.
- class brainvisa.data.fileSystemOntology.SetWeakAttr(*params)[source]¶
Bases:
SetAttrThis builder set the
ScannerRule.localAttributesattribute of the current rule.- Parameters:
params – list of attribute name followed by its value.
- build(scannerRule)[source]¶
- Parameters:
scannerRule – related
ScannerRule.
- brainvisa.data.fileSystemOntology.insert(path, *content)¶
Inserts rules in a
DirectoryScannerwhose pattern matches path.This function is only visible from inside a type or hierarchy file, during loading.
- brainvisa.data.fileSystemOntology.insertFirst(path, *content)¶
Inserts rules in a
DirectoryScannerwhose pattern matches path at the beginning of the list of rules.This function is only visible from inside a type or hierarchy file, during loading.
- brainvisa.data.fileSystemOntology.insertLast(path, *content)¶
Appends rules in a
DirectoryScannerwhose pattern matches path.This function is only visible from inside a type or hierarchy file, during loading.
brainvisa.data.sqlFSODatabase¶
This module contains classes defining Brainvisa databases.
The main classes are SQLDatabases and SQLDatabase.
- class brainvisa.data.sqlFSODatabase.Database[source]¶
Bases:
objectBase class for Brainvisa databases.
- findTransformationPaths(source_referential, destination_referential, maxLength=None, bidirectional=False)[source]¶
Return a generator object that iterate over all the transformation paths going from source_referential to destination_referential. A transformation path is a list of ( transformation uuid, destination referentia uuid). The pathsare returned in increasing length order. If maxlength is set to a non null positive value, it limits the size of the paths returned. Source and destination referentials must be given as string uuid.
- exception brainvisa.data.sqlFSODatabase.NotInDatabaseError[source]¶
Bases:
DatabaseError
- class brainvisa.data.sqlFSODatabase.SQLDatabase(sqlDatabaseFile, directory, fso=None, context=None, otherSqliteFiles=[], settings=None)[source]¶
Bases:
DatabaseA Brainvisa database with files stored in a hierarchically organized directory and a SQL database indexing the files according to Brainvisa ontology.
The SQL database is implemented using SQLite.
- checkTables()[source]¶
Checks if all types currently defined in the database ontology have a matching table in the sqlite database. It may be not the case when the database has been updated with a version of brainvisa that has not all the toolboxes. It should then be updated.
- findReferentialNeighbours(source_referential, cursor=None, bidirectional=True, flat_output=False)[source]¶
From one referential, find all referentials directly linked by transforms and return a tuple (referentials, paths), where paths is a dictionary which contains a list of transforms that leads to each referential (key of the dictionary) from the source_referential (a transform is a triplet (uuid_transform, uuid_from, uuid_to))
If flat_output is True, the output is a list of tuples (transform, source, dest).
- class brainvisa.data.sqlFSODatabase.SQLDatabases(databases=[])[source]¶
Bases:
DatabaseThis object stores several
SQLDatabaseobjects.- changeDiskItemFormatToSeries(diskItem)[source]¶
Changes the format of the diskItem to Series of diskItem.format The number is extracted from the name to begin the name_serie list attribute. Other files with the same name but another number are searched in the parent directory to find the other numbers of the serie.
brainvisa.data.readdiskitem¶
This module defines the class ReadDiskItem which is a subclass brainvisa.data.neuroData.Parameter.
It is used to define an input data file as a parameter in a brainvisa.processes.Process brainvisa.data.neuroData.Signature.
- class brainvisa.data.readdiskitem.ReadDiskItem(diskItemType, formats, requiredAttributes={}, enableConversion=True, ignoreAttributes=False, _debug=None, exactType=False, section=None, enableMultiResolution=False)[source]¶
Bases:
ParameterThe expected value for this parameter must be a readable
brainvisa.data.neuroDiskItems.DiskItem. This parameter type uses BrainVISA data organization to select possible files.- Syntax:
ReadDiskItem( file_type_name, formats [, required_attributes, enableConversion=1, ignoreAttributes=0 ]) formats <- format_name formats <- [ format_name, ... ] required_attributes <- { name : value, ...}
file_type_name enables to select files of a specific type, that is to say DiskItem objects whose type is either file_name_type or a derived type. The formats list gives the exhaustive list of accepted formats for this parameter. But if there are some converters ( see the section called “Role”) from other formats to one of the accepted formats, they will be accepted too because BrainVISA can automatically convert the parameter (if enableConversion value is 1, which is the default). Warning : the type and formats given in parameters of ReadDiskItem constructor must have been defined in BrainVISA types and hierarchies files. required_attributes enables to add some conditions on the parameter value : it will have to match the given attributes value.
This method of files selection ease file selection by showing the user only files that matches type and format required for this parameter. It also enables BrainVISA to automatically fill some parameters values. The ReadDiskItem class has methods to search matching diskitems in BrainVISA databases :
ReadDiskItem.findItems(<database directory diskitem>, <attributes>) : this method returns a list of diskitems that exist in that database and match type, format and required attributes of the parameter. It is possible to specify additional attributes in the method parameters. Found items will have the selected value for these attributes if they have the attribute, but these attributes are not mandatory. That’s the difference with the required attributes set in the constructor.ReadDiskItem.findValues(<value>): this method searches diskitems matching the value in parameter. This value can be a diskitem, a filename, a dictionary of attributes.ReadDiskItem.findValue(<value>): this method returns the best among possible value, that is to say with the more common attributes, highest priority. If there is an ambiguity, it returns None.
Examples
>>> ReadDiskItem( 'Volume 3D', [ 'GIS Image', 'VIDA image' ] ) >>> ReadDiskItem( 'Cortical folds graph', 'Graph', requiredAttributes = { 'labelled' : 'No', 'side' : 'left'} )
In the first example, the parameter will accept only a file whose type is 3D Volume and format is either GIS image or VIDA image, or a format that can be converted to GIS or VIDA image. These types and formats must have been defined first. In the second example, the parameter value type must be “Cortical folds graph”, its format must be “Graph”. The required attributes add some conditions : the graph isn’t labelled and represents the left hemisphere.
- checkValue(name, value)[source]¶
This functions check if the given value is valid for the parameter. If the value is not valid it raises an exception.
- property database¶
Returns the database this disk item belongs to
- diskItemDistance(diskItem, other)[source]¶
Returns a value that represents a sort of distance between two DiskItems. The distance is not a number but distances can be sorted.
- editor(parent, name, context)[source]¶
Virtual function that can be overriden in derived class. The function should return an object that can be used to edit the value of the parameter. This one raises an exception saying that no editor exist for this type.
- findValue(selection, requiredAttributes=None, preferExisting=False, ignore_db_formats_sorting=False, _debug=<undefined>)[source]¶
Find the best matching value for the ReadDiskItem, according to the given selection criterions.
The “best matching” criterion is the maximum number of common attributes with the selection, with required attributes satisfied.
In case of WriteDiskItem, if preferExisting, also search for value already in database.
If there is an ambiguity (no matches, or several equivalent matches), None is returned.
- Parameters:
selection (diskitem, or dictionary, or str (filename))
requiredAttributes (dictionary (optional))
preferExisting (boolean)
ignore_db_formats_sorting (boolean) – by default, in Axon >= 4.6.2, formats are sorted according to database-specific formats orders, thus overriding process-specified formats orders. This flag allows to disable this behavior.
_debug (file-like object (optional))
- Returns:
matching_value
- Return type:
DiskIteminstance, or None
- findValues(selection, requiredAttributes={}, write=False, _debug=<undefined>)[source]¶
Find all DiskItems matching the selection criterions
- listEditor(parent, name, context)[source]¶
Virtual function that can be overriden in derived class. The function should return an object that can be used to edit a list of values for the parameter. This one raises an exception saying that no list editor exist for this type.
- toolTipText(parameterName, documentation)[source]¶
Returns the text of a tooltip (in HTML format) for this parameter. Can be displayed as information in a GUI. The tooltip shows the name of the parameter, indicates if it is optional, and shows
documentationas a description of the parameter.
- typeInfo(translator=None)[source]¶
Returns a tuple containing (“Type”, type_name). The type_name is the name of the class possibly translated with the give translator or Brainvisa default translator if None.
- valueLinked(parameterized, name, value)[source]¶
This method is a callback called when the valueLinkedNotifier is activated. This notifier is shared between this parameter and the initial parameter in the static signature of the process. So when this function is called self is the initial parameter in the static signature. That why self is not used in this function.
brainvisa.data.writediskitem¶
This module defines the class WriteDiskItem which is a subclass brainvisa.data.neuroData.Parameter.
It is used to define an output data file as a parameter in a brainvisa.processes.Process brainvisa.data.neuroData.Signature.
- class brainvisa.data.writediskitem.WriteDiskItem(diskItemType, formats, requiredAttributes={}, exactType=False, ignoreAttributes=False, _debug=None, section=None)[source]¶
Bases:
ReadDiskItemThe expected value for this parameter must be a writable
brainvisa.data.neuroDiskItems.DiskItem.- Syntax:
WriteDiskItem( file_type_name, formats [, required_attributes={}, exactType=0, ignoreAttributes=0] ) formats <- format_name formats <- [ format_name, ... ]
This parameter type is very close to ReadDiskItem (WriteDiskItem derives from ReadDiskItem), but it accepts writable files. That is to say, it accepts not only files that are accepted by a ReadDiskItem but also files that doesn’t exist yet. It has the same search methods as the ReadDiskItem class but these methods generate diskitems that may not exist yet, using data ontology information.
brainvisa.data.actions¶
- class brainvisa.data.actions.FileProcess(file, action, pattern=None, diskItem=None)[source]¶
Bases:
objectRepresents a process to execute on a file. @type file: string @ivar file: the file to process @type action: Action @ivar action: the action to do to process the file @type selected: boolean @ivar selected: if the process is selected, the action will be executed by process method of a DBProcessor. @type pattern: regexp @ivar pattern: regular expression that describes the files to process in the directory self.file @type files: list of string @ivar files: if pattern is not None, files corresponding to that pattern that have been treated by doit method. useful for undoCmd.
- class brainvisa.data.actions.Move(dest, patternSrc=None, patternDest=None)[source]¶
Bases:
ActionAction to move src item to dest item. items can be files or directories. If intermediate directories in dest don’t exist they are created. Paths must be explicite, * cannot be used instead of file name. It is possible to rename at the same time : Examples : Move(destDir).doit(srcDir/item) : srcDir/item -> destDir/item (not renamed) Move(destDir, None, newItem).doit(srcDir/item) : srcDir/item -> destDir/newItem (renamed with fixed name) Move(destDir, “.*”, “-suffix”).doit(srcDir/item) : srcDir/item -> destDir/item-suffix (renamed with a name related to src name)
@type dest: string @param dest: directory where to move src @type patternSrc: string @param patternSrc: regular expression that will be matched on files to move @type patternDest: string @param patternDest: regular expression to expand the match object found on file to move in order to get the new name
brainvisa.data.databaseCheck¶
- class brainvisa.data.databaseCheck.BVChecker_3_1(db, context=None)[source]¶
Bases:
DBCheckerChecker for Brainvisa 3.1 hierarchy.
- process(component=None, debug=False)[source]¶
For each data in fileProcesses, calls associated action doit method which does the action to process the database. There is no possible undo for this processor. Calls process method for each components. @type component: string @param component: key of a component to process only a part of the database
- class brainvisa.data.databaseCheck.BVConverter_3_1(db, context=None, segmentDefaultDestination=None, grapheDefaultDestination=None)[source]¶
Bases:
DBConverterConvert database to Brainvisa 3.1 format. It contains a converter for each toolbox. To add a converter for a part of the database, create a sub class of DBConverter and redefine findActions method which goes throught the database and lists modification to do. Then, an instance of that converter must be added to this class’s components list
- findActions(component=None)[source]¶
Traiter les fichiers restants dans segment et graphe : les mettre dans la toolbox par defaut choisie -> ajouter l’option dans le process de conversion
- generateUndoScripts(component=None)[source]¶
Creates a script to undo conversion actions. The script file is dbDir/undoScriptName For each done action, the undoCmd is written in the undo script. Calls generateUndoScripts for each sub converter. @type component: string @param component: key of a component to process only a part of the database
- process(component=None, debug=False)[source]¶
For each stored actions, calls doit method which does the action to process the database. Each done action is stored in doneActions list in order to create undo scripts. Calls convert method for each sub converters. @type component: string @param component: key of a component to process only a part of the database
- class brainvisa.data.databaseCheck.DBChecker(db, context=None)[source]¶
Bases:
DBProcessorBase class for Database checker. This processor search some type of data in the database, checks them and eventually suggests an action to correct the data. @type db: Directory @ivar db: database directory @type filters: list @ivar filters: attributes whose values are used to sort and group data found in the database. @type searchTypes: list @ivar searchTypes: type of data searched for in the database @type fileProcesses: SortedDictionary (to keep insertion order) @ivar fileProcesses: map of data found in the database, grouped by filter attributes values. { filter1_value1 : {filter2_value1 : {… {filtern_value1: [ files ] }, filter1_value2 : { … } } }
- findActions(filters={}, component=None)[source]¶
Search for actions to do to process the database. @type component: string @param component: key of a component to process only a part of the database @rtype : list of FileProcess @return : actions to do to process the database
- process(component=None, debug=False)[source]¶
For each data in fileProcesses, calls associated action doit method which does the action to process the database. There is no possible undo for this processor. Calls process method for each components. @type component: string @param component: key of a component to process only a part of the database
- class brainvisa.data.databaseCheck.DBCleaner(db, context=None)[source]¶
Bases:
DBProcessorThis processor search unknown files in the database for brainvisa filesystem ontology. Default action proposed for these files is Remove. An unknown DiskItem in the database has _identified field false.
@type db : Directory @param db : diskitem that represents the database directory
- class brainvisa.data.databaseCheck.DBConverter(dbDir, context=None)[source]¶
Bases:
DBProcessorConverts a file system database by moving, renaming, removing files and directories. Generic converter, it can contain other converters, specialized for a part of the database.
- convertFiles(conversionPatterns, oldDir, newDir, baseDir, currentAcquisition='', currentAnalysis='', currentSession='', starReplace='acquisition')[source]¶
Files will be moved to destDir/currentAcquisition/currentAnalysis/currentSession and renamed according to patterns. If there is * patterns, the corresponding files will be moved to this path replacing starReplace by match string @type conversionPatterns: list of tuple (2-4) @param conversionPatterns: each tuple contain : source pattern, dest pattern and eventually a star pattern. the star pattern is the same as the source pattern with a variable in the name which represents an attribute (acquisition, analysis…) @type oldDir : string @param oldDir : directory where diffusion files are currently (diffusion) @type newDir: string @param newDir: directory where diffusion files will be after processing conversion. (diffusion/acquisition) @type destDir : string @param baseDir: base directory where the files must be put (generally diffusion) @type starReplace: string @param starReplace: indicates what attribute the match string in star pattern must replace in dest path.
- class brainvisa.data.databaseCheck.DBProcessor(dbDir, context=None)[source]¶
Bases:
object@type dbDir: string @ivar dbDir: database directory @type components : list of DBProcessor @ivar components: it is possible to have several processors, each specialized for one database part. @type fileProcesses: list of FileProcess @ivar fileProcesses: files that must be processed to process the database. @type doneProcesses: list of FileProcess @ivar doneProcesses: actions really done by process method in the inverse order of execution. It is stored to create the undo scripts. To undo, it is necessary to do inverse of each done action in inverse order. @type undoScriptName: string @ivar undoScriptName: name of the script that will be generated by generateUndoScripts method to undo done processing on the database.
- findActions(component=None)[source]¶
Search for actions to do to process the database. @type component: string @param component: key of a component to process only a part of the database @rtype : list of FileProcess @return : actions to do to process the database
- generateUndoScripts(component=None)[source]¶
Creates a script to undo conversion actions. The script file is dbDir/undoScriptName For each done action, the undoCmd is written in the undo script. Calls generateUndoScripts for each sub converter. @type component: string @param component: key of a component to process only a part of the database
- process(component=None, debug=False)[source]¶
For each stored actions, calls doit method which does the action to process the database. Each done action is stored in doneActions list in order to create undo scripts. Calls convert method for each sub converters. @type component: string @param component: key of a component to process only a part of the database
- class brainvisa.data.databaseCheck.DiffusionChecker(db, context=None)[source]¶
Bases:
DBCheckerChecker for t1mri toolbox. Checks data generated by the segmentation pipeline. All data must have a referential : each Raw T1 MRI have its own referential, each generated data have the same referential as corresponding raw t1 mri.
- class brainvisa.data.databaseCheck.DiffusionConverter(dbDir, context=None)[source]¶
Bases:
DBConverterAdd acquisition level in center/subject/diffusion if needed.
- class brainvisa.data.databaseCheck.PETConverter(dbDir, context=None)[source]¶
Bases:
DBConverterAdd acquisition level in center/subject/pet if needed. and analysis/segmentation and analysis/ROI
- class brainvisa.data.databaseCheck.T1MriChecker(db, context=None)[source]¶
Bases:
DBCheckerChecker for morphologist toolbox. Checks data generated by the segmentation pipeline. All data must have a referential : each Raw T1 MRI have its own referential, each generated data have the same referential as corresponding raw t1 mri.
- class brainvisa.data.databaseCheck.T1MriConverter(dbDir, context=None)[source]¶
Bases:
DBConverterIn protocol/subject :
protocol -> center
anatomy -> t1mri/acquisition/analysis (raw t1 mri and acpc coordinates files are put in acquisition, the others in analysis)
If anatomy already contains acquisition directories, they are moved in t1mri, and analysis level is added. * segment -> t1mri/acquisition/analysis/segmentation * tri, mesh -> t1mri/acquisition/analysis/segmentation/mesh * deepnuclei -> t1mri/acquisition/analysis/nuclei * Referential and transformations :
<subject>_TO_talairach.trm -> <center>/<subject>/registration/RawT1-<subject>_<acquisition>_TO_Talairach-ACPC.trm
<subject>.referential -> *<subject>-default_acquisition.referential, *<subject>_TO_.trm -> <subject>_default_acquisition_TO_.trm
- findActions()[source]¶
Search for actions to do to process the database. @type component: string @param component: key of a component to process only a part of the database @rtype : list of FileProcess @return : actions to do to process the database
- moveDir(src, dest, newName=None)[source]¶
Moves a directory in another directory adding levels acquisition and analysis. If src contains acquisition directories, they are moved to dest/acquisition/default_analysis/newName. if there is no acquisition level in src, directory containt is moved to dest/default_acquisition/default_analysis/newName.
- moveSessionGraphs(oldDir, newDir, destDir, subject)[source]¶
search for session directories in graph dir : a directory which doesn’t end with .data the session directory is renamed in session_auto and the graph files also @param oldDir : directory where the recognition sessions are during parsing @param newDir : directory where the recognition sessions are during conversion @param destDir : directory where the recognition sessions must be moved
brainvisa.data.directory_iterator¶
brainvisa.data.fileformats¶
brainvisa.data.ftpDirectory¶
- class brainvisa.data.ftpDirectory.FTPDirectory(name, server, login, password, baseDirectory='/')[source]¶
Bases:
VirtualDirectory- class Item(s, ftp, path)[source]¶
Bases:
Item- fullName()[source]¶
Return an identifier wich is unique in the VirtualDirectory where the data comes from. This identifier can be stored and used later with the VirtualDirectory’s get() method in order to get a VirtualDirectory.Item for the same data. May return None if the identifier cannot be build. A typical value for this identifier is the full path of a file relative to the virtual directory.
- name()[source]¶
Return the short name of the data. If not empty, this value can be used to create a file or directory name for the data. Therefore, it does not contain directory separator nor characters forbidden for file names. A typical value for name() is the file name without its parent path.
brainvisa.data.labelSelection¶
@author: Dominique Geffroy @organization: U{NeuroSpin<http://www.neurospin.org>} and U{IFR 49<http://www.ifr49.org>} @license: U{CeCILL version 2<http://www.cecill.info/licences/Licence_CeCILL_V2-en.html>}
- class brainvisa.data.labelSelection.LabelSelection(model=None, nomencl=None, **kwargs)[source]¶
Bases:
ParameterSelect labels in a nomenclature.
brainvisa.data.minfExtensions¶
Registration of all BrainVISA specific minf formats.
@author: Yann Cointepas @organization: U{NeuroSpin<http://www.neurospin.org>} and U{IFR 49<http://www.ifr49.org>} @license: U{CeCILL version 2<http://www.cecill.info/licences/Licence_CeCILL_V2-en.html>}
brainvisa.data.patterns¶
- class brainvisa.data.patterns.DictPattern(pattern)[source]¶
Bases:
objectA DictPattern contains a pattern that is matched against a string and a set of attributes contained in a dictionary. When the match succeed, it returns a dictionary containing attributes values extracted from the input string.
Such patterns are used to define Brainvisa ontology rules which associate filenames and data types.
The input pattern is a string that is splitted in three kinds of tokens:
An attribute name enclosed in
<and>A named regular expression enclosed in
{and}A string literal which is everything not enclosed neither in braces nor with
<and>.
When
DictPattern.match( s, dict )is called, all attribute names from the pattern are replaced by the corresponding value in dict. If the dict does not contain the attribute, the match fails. Then, the string is matched against the pattern. If the pattern contains named regular expressions, the values corresponding to each expression is put in the resulting dictionary. If the match succeed, a dictionary (possibly empty) is returned. Otherwise,Noneis returned.In the string literal of the pattern, special characters can be found:
*matches any string and the matched value is associated to afilename_variablekey in the results dictionary.#matches any number and the matches value is associated to aname_seriekey in the results dictionary.
- Match examples:
p = DictPattern( '<subject>_t1' ) p.match( 's_t1', { 'subject': 's' } ) == {} p.match( 's_t1', { 'subject': 'x' } ) == None p.match( 's_t2', { 'subject': 's' } ) == None p = DictPattern( '{subject}_t1' ) p.match( 's_t1', { 'subject': 's' } ) == {'subject': 's'} p.match( 'tutu_t1', { 'subject': 's' } ) == {'subject': 'tutu'} p.match( 's_t2', { 'subject': 's' } ) == None p = DictPattern( '*_#' ) p.match( 'toto_titi', {} ) == None p.match( 'toto_42', {} ) == {'name_serie':'42', 'filename_variable':'toto'} p = DictPattern( 'begin*_<subject>_*end' ) p.match( 'beginxxx_anatole_yyyend', { 'subject': 'anatole' } ) == None p.match( 'beginxxx_anatole_xxxend', { 'subject': 'anatole' } ) == { 'filename_variable': 'xxx' } p.match( 'beginxxx_anatole_xxxend', { 'subject': 'raymond' } ) == None p = DictPattern( 'begin#_<subject>_#end': ( p.match( 'beginxxx_anatole_xxxend', { 'subject': 'anatole' } ) == None p.match( 'begin123_anatole_456end', { 'subject': 'anatole' } ) == None p.match( 'begin123_anatole_123end', { 'subject': 'anatole' } ) == { 'name_serie': '123' } p.match( 'begin123_anatole_123end', { 'subject': 'raymond' } ) == None
- Unmatch example:
>>> DictPattern.unmatch( matchResult, dict )
This allows to build the string that would produce matchResult if
DictPattern.match( s, dict )is succesfully used. The unmatch always succeed if matchResult is not None, in this case, we have :>>> DictPattern.match( DictPattern.match( s, dict ), dict ) == s
- match(s, dict)[source]¶
Checks if the given string matches the pattern.
- Parameters:
s (string) – the string which should match the pattern
dict – a dictionary containing the values to set to the attributes named in the pattern.
- Returns:
a dictionary containing the value of each named expression of the pattern found in the string, None if the string doesn’t match the pattern.
- unmatch(matchResult, dic)[source]¶
The opposite of
match()method: the matching string is found from a match result and a dictionary of attributes values.- Parameters:
matchResult – dictionary which associates a value to each named expression of the pattern.
dict – dictionary which associates a value to each attribute name of the pattern.
- Return type:
string
- Returns:
the rebuilt matching string.
brainvisa.data.sql¶
brainvisa.data.temporary¶
- class brainvisa.data.temporary.TemporaryFileManager(directory, defaultPrefix)[source]¶
Bases:
objectThis object manages temporary files. It enables to create new temporary files that will be automatically deleted when there is no more references on them.
- new(suffix=None, prefix=None, directory=None)[source]¶
Creates a new temporary file. The filename will be directory/prefix+pid+count+suffix
- Parameters:
suffix (string) – something to add at the end of the generated filename.
prefix (string) – something to add at the begining of the filename. A default prefix is used if None.
directory (string) – path of the directory where the file must be created. A default directory is used if None.
- Returns:
an internal object that contains the filename and enables to destroy the file when it is no more used.
brainvisa.data.virtualDirectory¶
- class brainvisa.data.virtualDirectory.VirtualDirectory(manager=None)[source]¶
Bases:
objectA VirtualDirectory is a place where file- or directory-like data are stored (local file system, ftp server, data base, etc.). A VirtualDirectory have a name. That name is used whith a VirtualDirectoriesManager to build URL-like identifiers.
- class Item(virtualDirectory=None)[source]¶
Bases:
UserDictA VirtualDirectory.Item is either a binary block of data or directory like structure containing VirtualDirectory.Items.
- fullName()[source]¶
Return an identifier wich is unique in the VirtualDirectory where the data comes from. This identifier can be stored and used later with the VirtualDirectory’s get() method in order to get a VirtualDirectory.Item for the same data. May return None if the identifier cannot be build. A typical value for this identifier is the full path of a file relative to the virtual directory.
- globalName()[source]¶
Return an identifier wich is unique in the VirtualDirectoryManager where the VirtualDirectory containing the data is registered. This identifier can be stored and used later with the virtual directory manager’s get() method in order to get a VirtualDirectory.Item for the same data. May return an empty string if the identifier cannot be build.
- name()[source]¶
Return the short name of the data. If not empty, this value can be used to create a file or directory name for the data. Therefore, it does not contain directory separator nor characters forbidden for file names. A typical value for name() is the file name without its parent path.
- parent()[source]¶
Return the virtualDirectory.Item corresponding to this item’s parent directory. May return None if either the parent cannot be easily found (e.g. in a non hierarchical storage implementation).