Note
Click here to download the full example code
Graph building¶
Creating a complete graph and nomenclature in Python
from __future__ import print_function
from __future__ import absolute_import
import anatomist.direct.api as ana
from soma import aims
from soma.qt_gui.qt_backend import Qt
import sys
# create graph structure
graph = aims.Graph('RoiArg')
# these are needed for internal conversions
graph['type.global.tri'] = 'roi.global.tri'
graph['roi.global.tri'] = 'roi roi_global.gii aims_mesh'
# create 2 nodes
v = graph.addVertex('roi')
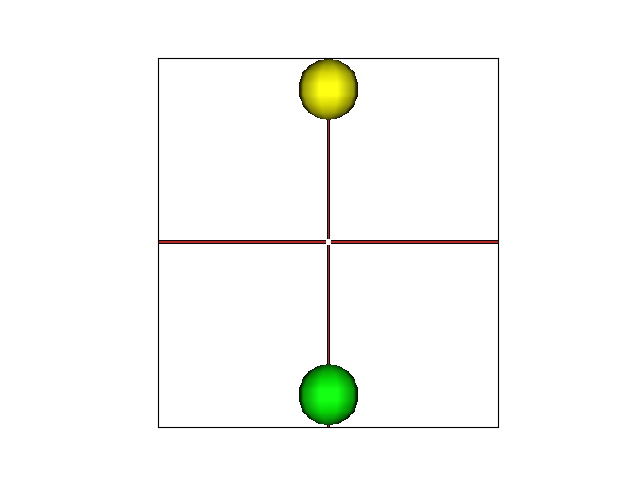
mesh = aims.SurfaceGenerator.sphere([0, 0, 0], 10, 100)
aims.GraphManip.storeAims(graph, v, 'aims_mesh', mesh)
v['name'] = 'sphere1'
v = graph.addVertex('roi')
mesh = aims.SurfaceGenerator.sphere([0, 0, 100], 10, 100)
aims.GraphManip.storeAims(graph, v, 'aims_mesh', mesh)
v['name'] = 'sphere2'
# create a corresponding nomenclature
hie = aims.Hierarchy()
hie.setSyntax('hierarchy')
hie['graph_syntax'] = 'RoiArg'
n = aims.Tree(True, 'fold_name')
n['name'] = 'sphere1'
n['color'] = aims.vector_S32([255, 255, 0])
hie.insert(n)
n = aims.Tree(True, 'fold_name')
n['name'] = 'sphere2'
n['color'] = aims.vector_S32([0, 255, 0])
hie.insert(n)
runloop = Qt.QApplication.instance() is not None
# create anatomist objects
a = ana.Anatomist()
ahie = a.toAObject(hie)
ahie.releaseAppRef()
agraph = a.toAObject(graph)
agraph.releaseAppRef()
# display graph
w = a.createWindow('3D')
w.addObjects(agraph, add_children=True)
br = a.createWindow('Browser')
br.addObjects(ahie)
w.sphinx_gallery_snapshot()
if runloop and 'sphinx_gallery' not in sys.modules:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del w, br, agraph, ahie, v, graph, hie, n
Total running time of the script: ( 0 minutes 0.453 seconds)