Note
Click here to download the full example code
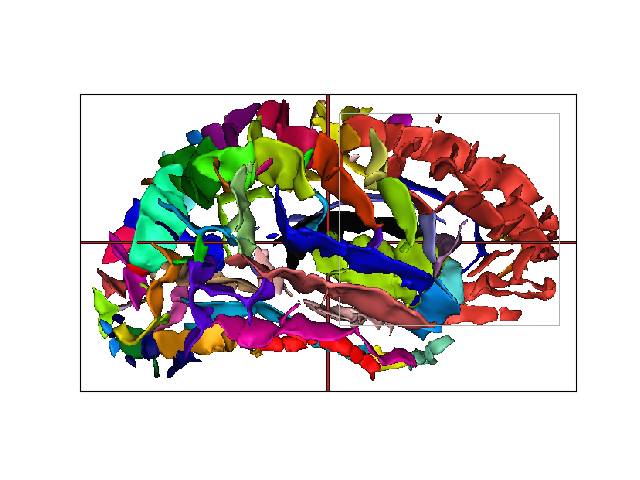
Selection by nomenclature¶
Selecting graph nodes according a nomenclature
from __future__ import absolute_import
import anatomist.direct.api as anatomist
from soma import aims
from soma.qt_gui.qt_backend import Qt
import os
import sys
runloop = Qt.QApplication.instance() is None
a = anatomist.Anatomist()
sh = aims.carto.Paths.shfjShared()
nom = a.loadObject(os.path.join(sh, 'nomenclature', 'hierarchy',
'sulcal_root_colors.hie'))
graph = a.loadObject('Rbase.arg')
w = a.createWindow('3D')
w.addObjects(graph, add_graph_nodes=True)
a.execute('SelectByNomenclature', names='PREFRONTAL_right', nomenclature=nom)
# to unselect all
# a.execute( 'Select' )
if 'sphinx_gallery' in sys.modules:
# display in matplotlib for sphinx_gallery
w.camera(view_quaternion=[0.5, -0.5, -0.5, 0.5])
w.windowConfig(view_size=[642, 384])
w.sphinx_gallery_snapshot()
runloop = False
if runloop:
Qt.QApplication.instance().exec_()
if runloop or 'sphinx_gallery' in sys.modules:
del w, graph, nom
Total running time of the script: ( 0 minutes 2.512 seconds)