Restricted access applications¶
Constellation: Connectivity-based Parcellation¶
Cortex parcellation based on structural connectivity information. It performs group-wise parcellations, either on a “group-only” basis (one group result), or provides individual results, from a group analysis. The whole cortex is parcelled by patches.
This toolbox, developed in NeuroSpin / UNATI lab, will be released “soon” when it is completely ready.
Gingko: Diffusion MRI and connectivity¶
Diffusion imaging processing, tractography and bundles analysis.
Gingko (formerly Connectomist-2) was developed by The Gingko lab of NeuroSpin (Cyril Poupon), in a different software framework, it is also using Anatomist for all visualization needs. It features dMRI preprocessings (outliers detection, distortion correction), local diffusion models processing, tractography, and fibers bundles post-processing (bundles measurements, clustering…).
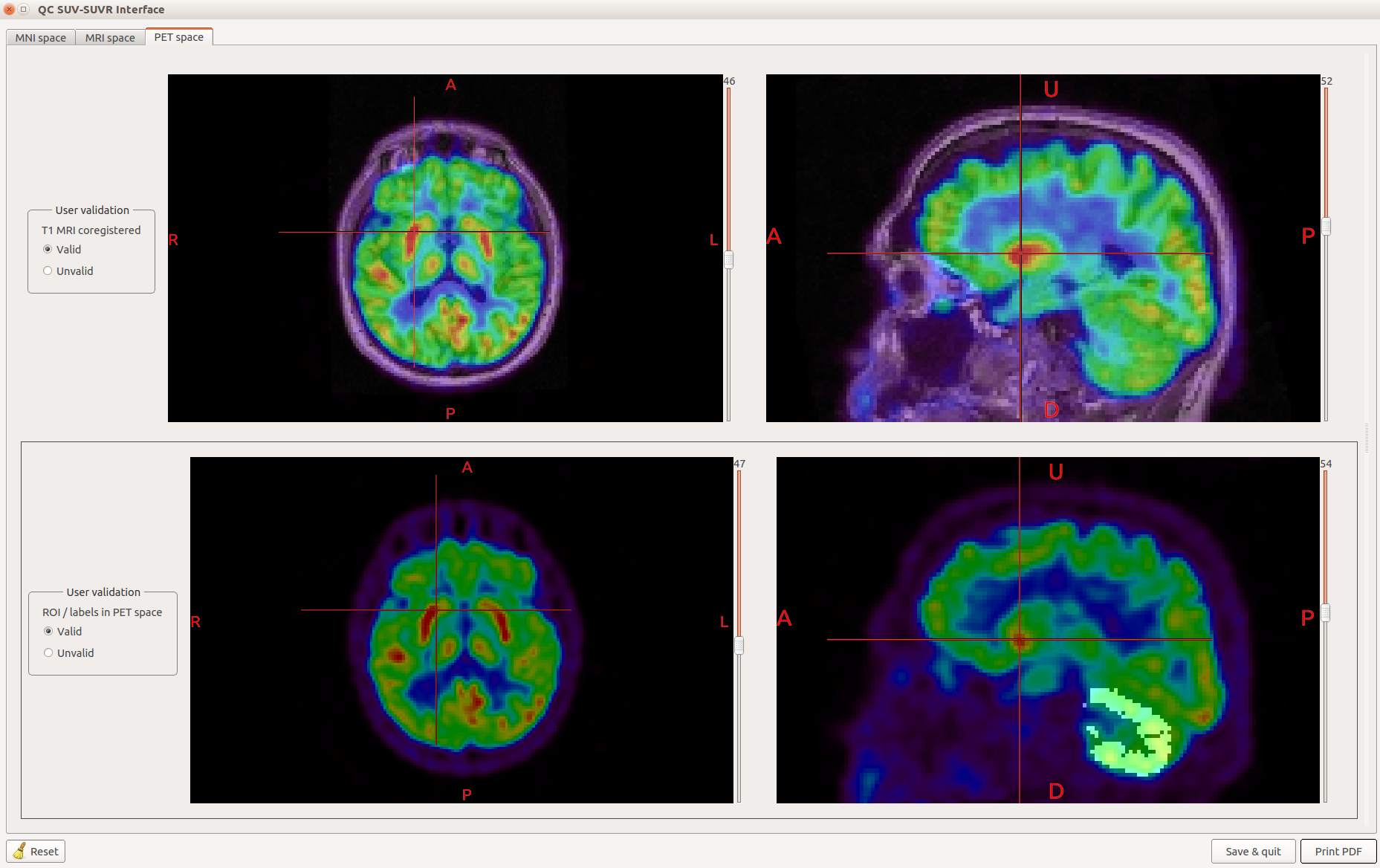
Nuclear imaging toolbox¶
A toolbox for BrainVISA developed by the  CATI project labs
CATI project labs
Baby toolbox¶
Semi-automatic segmentation toolbox for T2-weighted neonatal MRI images. This toolbox has been developed in Neurospin in the Developing Brain team. It is derived from Morphologist with adaptations to take into account the T2-weighted image contrast, the partially myelinated nature of baby brains, thus involving some steps of manual corrections, alterning automatic and interactive parts.
The toolbox is used internally and has no public release up to now.
SACHA: Hippocampus segmentation and morphometry¶

Automatic segmentation of hippocampus and amygdalia. Developed by the Aramis team, ICM, Paris.
WHASA: White matter hyper-intensities segmentation¶
Developed by the Aramis team, ICM, Paris.