What can you find in BrainVISA ?¶
BrainVISA provides a complete, modular, infrastructure for neuroimaging software. It helps organizing heterogeneous software and data and provides a common general graphical interface for users.
BrainVISA is thus a set of tools rather than a single software.
Axon is the name of the software which runs when you use the brainvisa command. The latter is still named brainvisa for historical reasons. It is a central software in the BrainVISA environment.
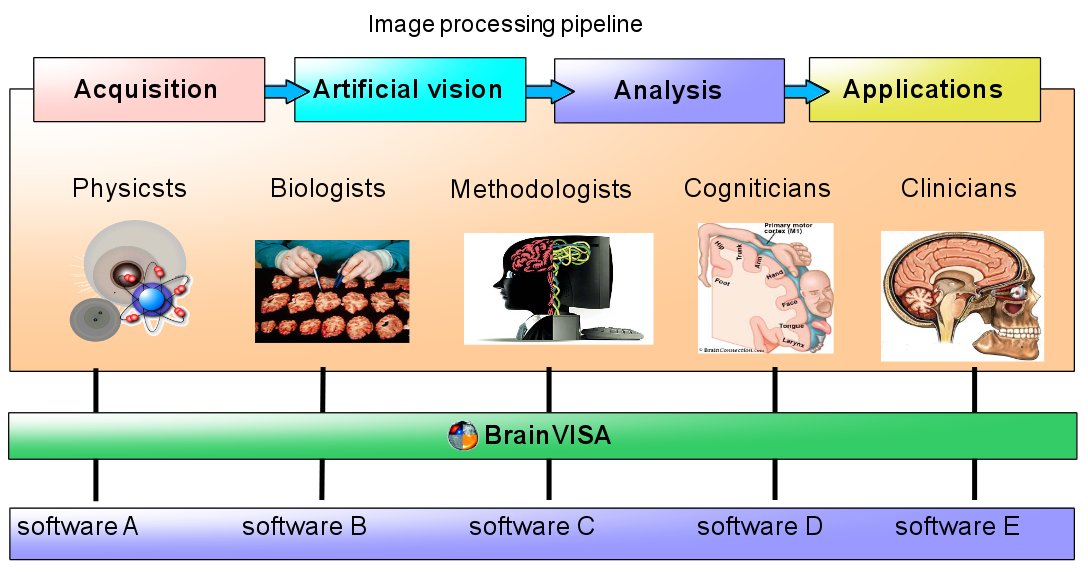
BrainVISA components include several aspects:
Data organization as directory trees with database indexing and querying
Interactive 3D visualization for all kind of neuroimaging data, supporting a wide range of data file formats: Anatomist: Data visualization and library of high-level neuroimaging graphical components
Software integration tools allowing to develop or interface new processing tools
Lower-level neuroimaging software libraries (AIMS) useable as building blocks for new processing tools
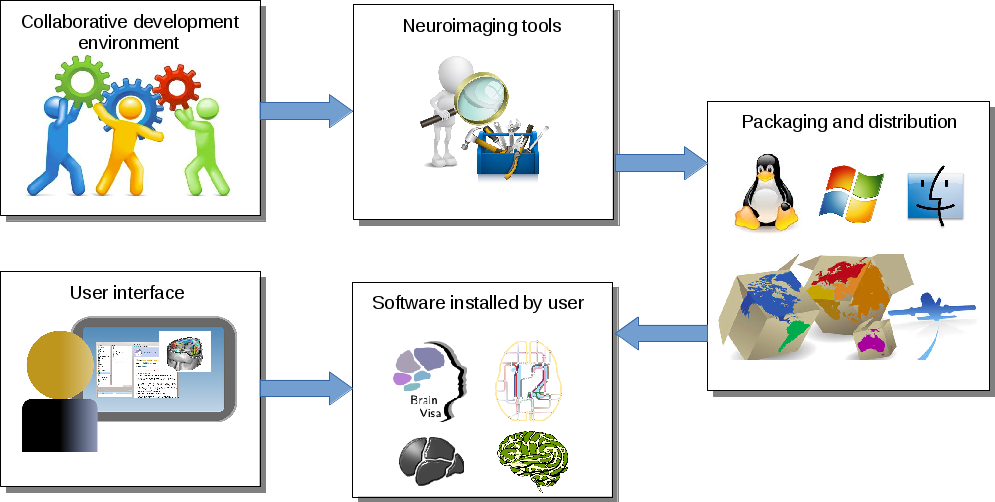
Axon¶
Axon provides a graphical user environment, and integrates the different tools and data to make them available for end users.
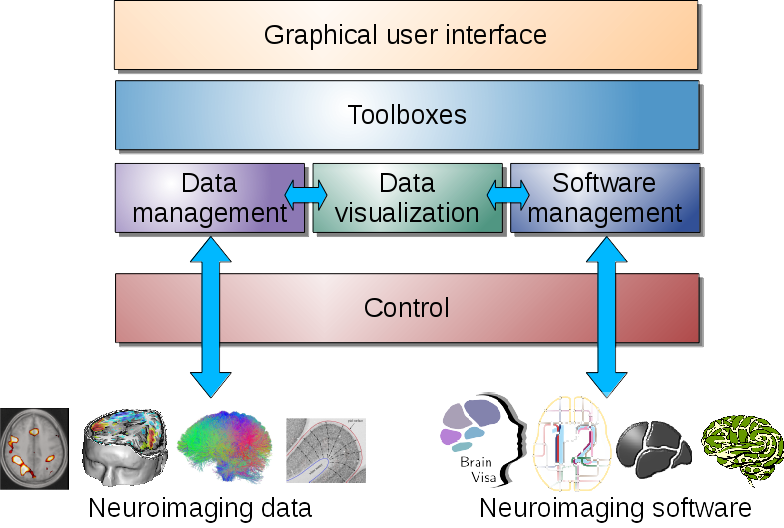
Unified graphical interface to data processing¶
Axon organizes processing tools and provides a unified graphical user interface for them.
This user interfaces are automatically generated by Axon from processes parameters characteristics.
However custom user interfaces are also possible and used in some applications, such as Morphologist UI.
Data management¶
Data is kept in organized directories, and referenced in databases. Files organization can be described to adapt a particular schema, for instance to allow reusing data from external sofftware such as Freesurfer or SPM. Databasing allows to query and filter data to be processed, and to generate processed file names.
In addition to files organization, Axon offers a way of automatically handling the complex problem of coordinates systems used by various data.
See The documentation for more details.
Software management¶
Different software can be interfaced with BrainVISA and Axon, even when they are using different languages and different conventions, coordinates systems, data organization…
Several BrainVISA toolboxes are already interfacing some popular software from the neuroimaging community, at least part of their tools: FreeSurfer, SPM, FSL… Other tools and software may be interfaced the same way via extensions.
Many software libraries are directly useable from BrainVISA toolboxes and processing tools, amongst which several external Python language libraries which prove very useful for the neuroimaging community: for instance Numpy, Scipy, Matplotlib, and many others.
Visualization¶
Visualization and interactive edition of data is integrated in Axon user interface, via the eye  and pencil
and pencil  icons close to data parameters in processing interfaces.
icons close to data parameters in processing interfaces.
Axon provides facilities to customize and write new specialized visualization tools. Most of them make use of Anatomist: Data visualization and library of high-level neuroimaging graphical components. Visualization can be adapted to specific needs for specific data, and complete 3D scenes can be built and triggered by a simple click.
Moreover coordinates systems are transparently and automatically managed by the combination of Axon databasing and Anatomist visualization capabilities.
Processing control infrastructure¶
Axon provides a processing pipeline management infrastructure: processes are organized with dependencies, allowing parallel execution.
Parallel or distributed execution may be performed either locally (on a mutiprocessor machine for instance), or remotely on a computing cluster. Distributed processing is managed via Soma-Workflow. This software library handles jobs dependencies and distribution, and interfaces in a transparent way remote computing clusters using various batch systems, or a local machine.
Integrate your tools¶

BrainVISA is designed to be customizable and extensible. User-defined research tools can be easily added either for personal convenience, for sharing research tools in a lab, or to distribute finalized toolboxed to the community.
All infrastructure software is free and works on Linux, Mac OS and Windows. Some additional toolboxes may contain non-free code and/or be limited to fewer platforms, but are out of topic here.
Several levels of integration may be reached.
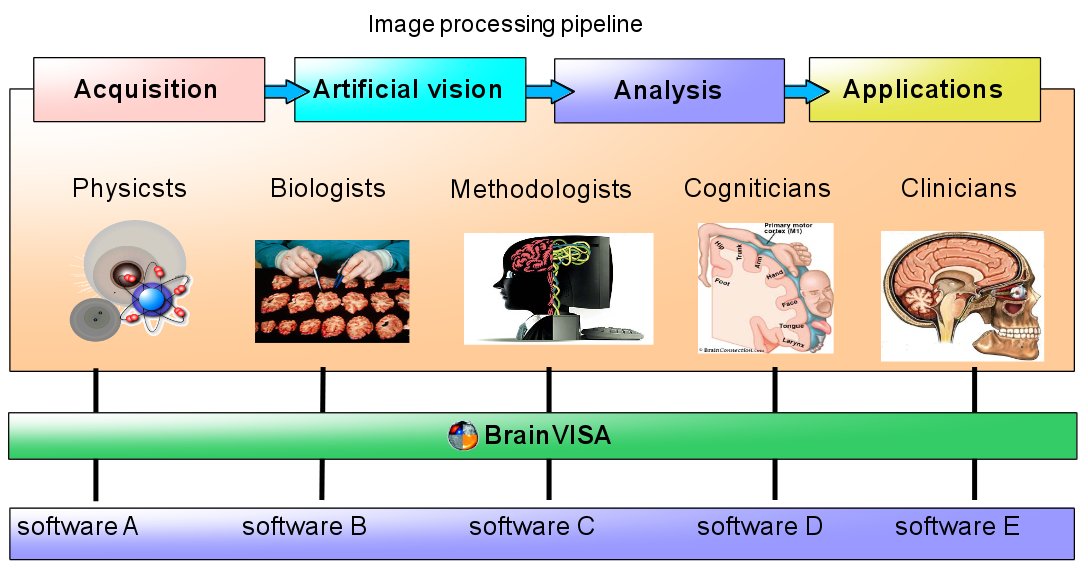
The levels correspond to different layers of the infrastructure, which may be schematically represented this way:
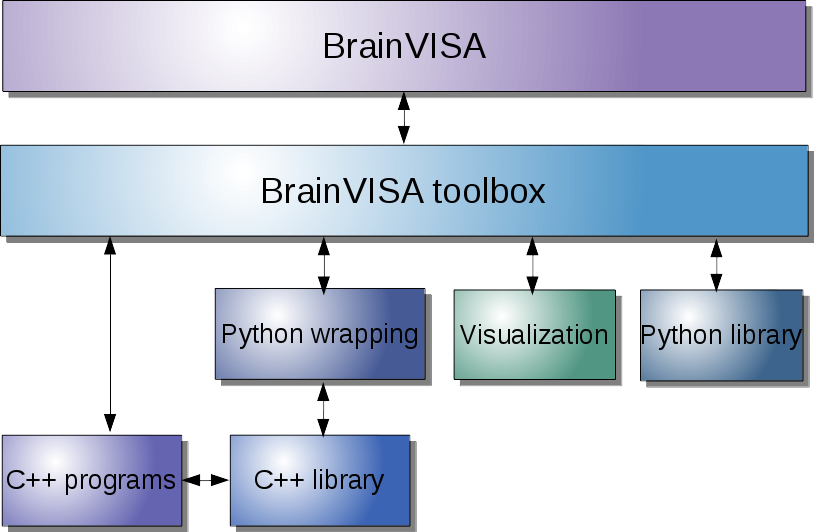
More details may be found in the following sections, which may become very technical.

Modularity¶
BrainVISA is a modular environment. It is possible to install, use, and build on a subset of the tools. Of course there are dependencies between elements but large parts are optional.
For instance we can cite a few typical use cases:
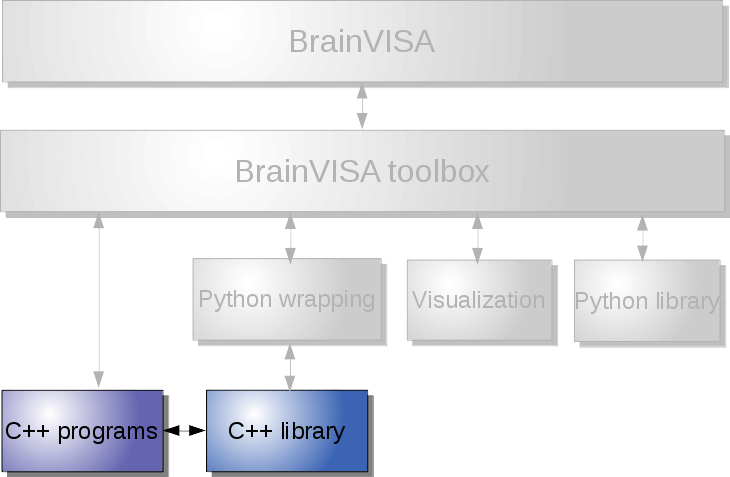
Low-level: use image processing commands and C++ libraries¶
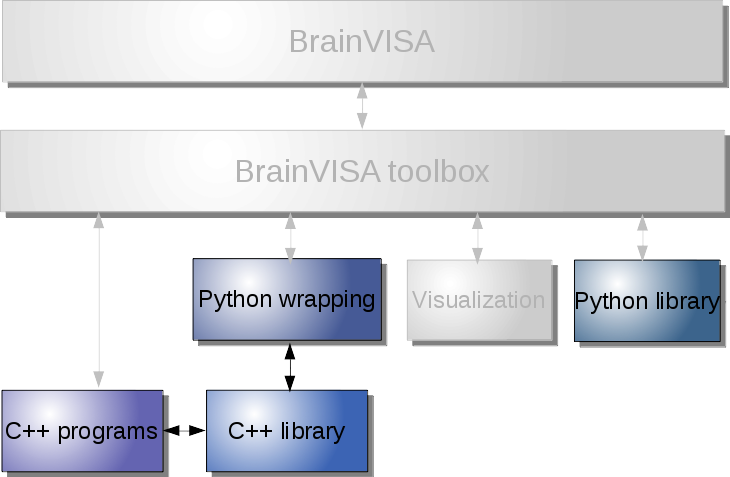
Image processing, with Python scripting¶
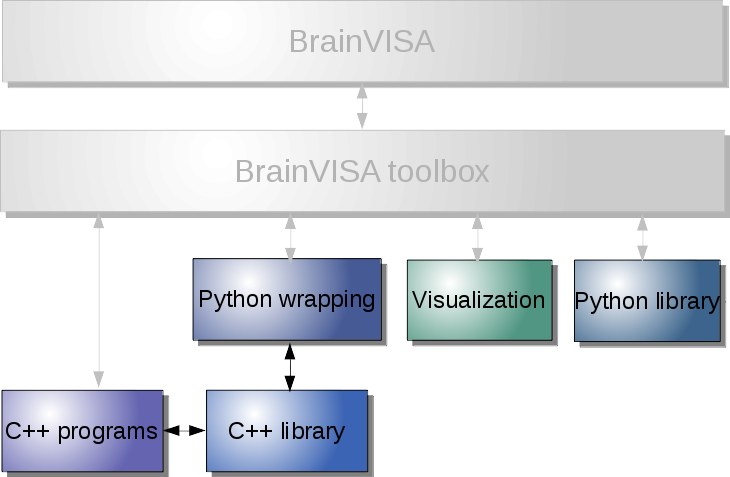
Visualization only¶
Axon, without visualization or builtin processing toolboxes¶
Adding personal processes to your BrainVISA¶
Adding processes may be done easily using a standard binary installation of BrainVISA: no need to access the source code of projects and to compile anything.
Processes may be added in a personal directory which is seen as a regular toolbox. Thus adding a personal process is just a matter of adding a Python script file in this processes directory.
The Programming course slides with corresponding data are a good starting point.
This is done within the Axon framework, using its programming interface.

Designing toolboxes¶
Toolboxes are a set of organized Axon processes, and may also contain types and files oprganization definitions, and documentation. They are also described in the Programming course slides.

Developing image analysis algorithms¶
BrainVISA provides image analysis libraries, which are mostly focused on anatomical MRI processing but may be used in various applications. They handle various types of data used in the field of neuroimaging, and unserstand a rather large set of file formats. These are the AIMS libraries. They include a set of C++ language libraries, some Python language bindings which make them scriptable in an easy way, and executable programs which may be direcly called by users or by scripts.
Programming in C++ allows lower-level, optimized image processing algorithms to be written. However this is at the cost of requiring compilation. There is a new tool to help developing and distributing tools: see Casa-distro. This tool allows to get libraries and compilation tools in Docker containers, so that they can be compiled and redistributed in an environment which is compatible with BrainVisa public binary distributions.
At lower level, the libraries are not available as binary installable files on every system, developing for compilation generally requires to compile the AIMS libraries from sources. Thus you will have to look at the sources repository section and the build system section.
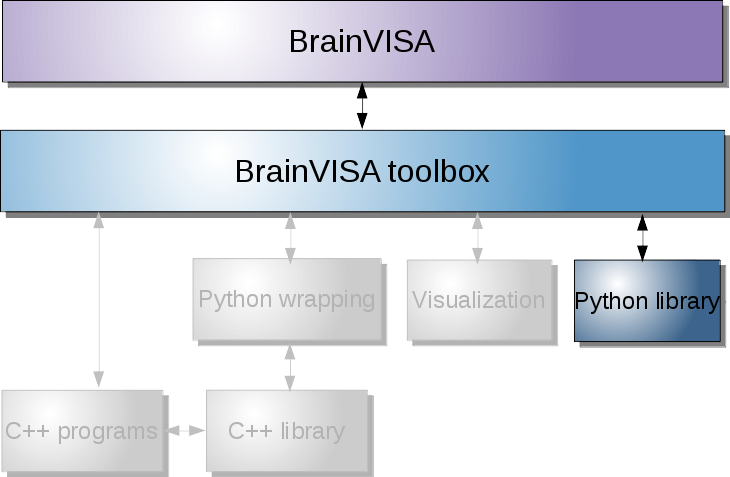
Providing custom interactive visualization¶
Visualization in BrainVISA can be provided via Axon through viewer processes. Most of them are making use of Anatomist for its interactive 3D capabilities, and its easy scripting facilities: PyAnatomist for scripting in Python language.

Distributing tools to the community¶
Tools based on BrainVISA development libraries can be freely distributed to the community. Distribution may be done via software packages which can be installed on top of existing BrainVISA installations, just as in the download section.
From version 4.5, BrainVISA is installed via a modular installer tool, similar to what is commonly found to manage software packages in a Linux distribution, but which works on all supported architectures (Linux, Mac, Windows), and allowing local installations: not needed administrator priviledges, and multiple versions of BrainVISA may be installed independently in different user directories.
It is possible to build custom packages in custom repositories, which can be added to the installer.
Details are available in the Packaging and installation system section.

Licensing policy¶
Toolboxes developers have some degrees of freedom on the licenses under which they can distribute their software. There are a few limitations, however:
Python graphical interfaces are GPL (CeCILL-v2): these graphical parts are built using PyQt, which is bound to GPL, and thus require all software built on them to be also distributed under a GPL-compatible licence. This applies to all Axon processes, which may appear in the brainvisa GUI.
But lower-level processing tools do not normally require such graphical interface, so are not bound to a specific license. Toolbox designers are thus free to choose the distribution license they prefer, provided their toolbox is separated in two distinct parts: the processing part (non-GUI, any license may be chosen), and the Axon part (GPL or CeCILL-v2).
Anatomist also has Qt user interface, but is globally built in C++ so does not have a strong dependency on PyQt: Anatomist is distributed under the CeCILL-B license (BSD-like). It has plugins in Python language, which may make use of PyQt however.
PyAnatomist, and python plugins, are using PyQt, so is distributed under the CeCILL-v2 (GPL-compatible) license. Custom anatomist plugins in Python must thus follow this licensing policy.
Technical developers section¶

Development environment¶
A new tool has been designed to setup very quickly a new developer into a situation when he can start writing, building and testing tools, without the burden of setting up a complete development system: this is Casa-distro. It will also ensure to build toolboxes and new tools that are compatible with public distributions of BrainVisa.
BrainVISA main software and programming libraries¶
All of the software here is free and portable on Linux, Mac and Windows. Some additional toolboxes may contain non-free code and/or be limited to fewer platforms, but are not listed here.
 Axon, conductor software:
Axon, conductor software:the “brainvisa” program and libraries, pipelining infrastructure, databasing, and graphical interface. Language: Python
 Anatomist 3D interactive visualization:
Anatomist 3D interactive visualization: Distributed processing: Soma-Workflow:
Distributed processing: Soma-Workflow:Language: Python
- AIMS image processing:
- Soma-base:
General python tools.
 CAPSUL pipelining system:
CAPSUL pipelining system:meant to replace the one currently in Axon. Language: Python
- Casa-Distro development and distribution environment:
development setup in Docker machines with packaging and distribution facilities.
- Brainvisa-Cmake build environment:
Language: Python, CMake
- |installer_head|:
Language: Python, CMake

Sources forge and repositories¶
Two repositories are used to host the source code of BrainVISA project. All components with public source code are on GitHub (either on BrainVISA organization or on Populse organization). Project with restricted read access are hosted on BioProj, the historical BrainVISA RedMine server. BioProj is operated by the CEA institution, and has access rules that forbids anonymous access. A login and password are mandatory to access to projects on BioProj. However there is a “public” login/password to access the open-source code that is still on BioProj (migration to GitHub is ongoing):
login: brainvisa
password: Soma2009
So it is normally possible to access the repository using this kind of link:
https://brainvisa:Soma2009@bioproj.cea.fr/neurosvn/brainvisa/
https://brainvisa:Soma2009@bioproj.cea.fr/redmine/projects/brainvisa
A GitHub project also holds some developers documentation: https://github.com/brainvisa/brainvisa.github.io

Build system¶
Projects are mainly in C++ and Python language. As BrainVISA is modular, it is made of multiple projects, which are separated by scope, aim, maintainers, language, and licence. To build the whole thing, we needed a tool to build an aggregation of the various projects. This tool is based on CMake but needed a higher-level tool to glue the various projects. This is the role of the brainvisa-cmake project of BrainVisa, and its main program, bv_maker.
Instructions on how to use it are available on the sources server web interface:
The building tools integrate software source repositories management, configuration, building, documentation generation, automatic testing, and software packaging. They thus allow to be used as a complete integrated development process.
As an example we have them running automatically every night on 7 different systems (Windows, Mac, and a set of Linux variants, including both virtual and physical machines). Problems are notified to the maintainers.

Packaging and installation system¶
The packaging / installation software is documented here: :installer:`BrainVISA Installer <index.html>`.
It allows to generate a packages repository, and the installer program available in the download section.
Brainvisa-installer can also be used via bv_maker which is generally mmore convenient since it allows to store all configuration and options in a config file.

Other BrainVISA libraries¶
Morphoplogist CAPSUL processes APIs: Morphologist processes constituting the main Morphologist pipeline have been converted to the new CAPSUL API.